Peptide loading complex
The peptide-loading complex (PLC)[1] is a short-lived, multisubunit membrane protein complex that is located in the endoplasmic reticulum (ER). It orchestrates peptide translocation and selection by major histocompatibility complex class I (MHC-I) molecules. Stable peptide-MHC I complexes are released to the cell surface to promote T-cell response against malignant or infected cells. In turn, T-cells recognize the activated peptides, which could be immunogenic or non-immunogenic.
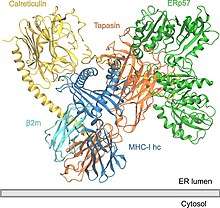
Overview
A PLC assembly consists of seven subunits, including the transporters associated with antigen processing (TAP1 and TAP2 – jointly referred to as TAP), the oxidoreductase ERp57, the MHC-I heterodimer, and the chaperones tapasin and calreticulin. TAP transports proteasomal degradation products from the cytosol into the lumen of the ER, where they are loaded onto MHC-I molecules. The peptide-MHC-I complexes then move via a secretory pathway to the cell surface, presenting their antigenic load to cytotoxic T-cells.
In general, preliminary MHC-I heavy chains are chaperoned by the calnexin–calreticulin system in the ER. Together with β2-microglobulin (β2m), MHC-I heavy chains form assemblies of heterodimers that act as receptors for antigenic peptides. Empty MHC-I heterodimers are recruited by calreticulin and form short-lived macromolecular PLC where the chaperone tapasin further provides stabilization in the MHC-I molecules. Furthermore, ERp57 and tapasin form disulfide-linked conjugates, and tapasin is crucial for maintaining the structural stability of the PLC as well as facilitating optimal peptide loading. After final quality control, during which MHC-I heterodimers undergo peptide editing, stable peptide–MHC-I complexes are released to the cell surface for T-cell recognition. The PLC can serve a large variety of MHC-I allomorphs, thus playing a central role in the differentiation and priming of T lymphocytes, and in controlling viral infections and tumour development.
Structure
So far, only the structure of human PLC has been elucidated using single-particle electron cryo-microscopy (cryo-EM). The structural organization consists of tapasin, calreticulin, ERp57, and MHC-I molecules that are centered around TAP, bearing a pseudo-symmetric orientation. PLC size is 150 Å by 150 Å with a total height of 240 Å across the ER membrane. Two opposing tapasin molecules shape the central domain in the PLC. In this model, residue E225 of the N-terminal immunoglobulin-like domain of one molecule and R60, which is located in the short helical motif of the seven-stranded N-terminal β-barrel of the second molecule, in salt-bridge distance. These residues are conserved among jawed vertebrates, but missing in avian PLC. This is consistent with the existence of a single copy of tapasin in avian PLC and the absence of an N-terminal transmembrane domain (TMD0) in avian TAP1. The ER-luminal domains of the opposing tapasin molecules are tilted 30° towards each other, positioning the two membrane entry points of the transmembrane helices 60 Å apart. The resulting domain twist excludes binding of calreticulin or calnexin to the glycan of the dimeric tapasin scaffold and explains why only mature tapasin can assemble into the PLC. Another structural feature is that ERp57 in its typical U-shaped conformation is complexed to tapasin via the catalytically active a and a′ domains. The unexpected orientation of tapasin enables the C-terminal extension of the a′ domain of the trans ERp57 to interact with the C-terminal immunoglobulin-like domain of the cis tapasin, potentially adding to the stability of the PLC.
This cryo-EM structure reveals a high degree of plasticity of the N-terminal three β-sheet sandwich of tapasin and shows that flexible fastening of MHC-I is essential for tapasin to exert its proofreading function. Calreticulin is crucial and is highly dependant in the assembly and maturation of MHC-I in the PLC. The globular lectin domain of calreticulin harbours a glycan-binding site, which senses monoglucose moieties of N-core glycosylated MHC-I before it associates with tapasin. The monoglucosylated branch of the N-core glycan that emanates from N86 of MHC-I is binding to the glycan-binding surface of calreticulin, whereas another mannose branch is likely to lie close among residues at the edge of the lectin β-sandwich. Consistent with its proposed calcium-dependent lipid-sensing activity, the C-terminal acidic tail of calreticulin points towards the ER-luminal membrane leaflet, where it is located close to the C-terminal immunoglobulin-like domain of tapasin. In the cryo-EM structure emphasis is given in the central role of the C-terminal immunoglobulin-like domain of tapasin, which acts as an essential multivalent interaction core for the remaining PLC subunits. A central cavity that connects the exit of the peptide translocation pathway of TAP with the ER lumen via two lateral windows might serve as a molecular basket for transported peptides before they are edited by ER-resident aminopeptidases.
TAP
TAP is a heterodimeric complex, consisting of TAP1 (ABCB2) and TAP2 (ABCB3) members of the ABC transporter superfamily. The common feature of all ABC transporters is their organization: 1) into two transmembrane domains (TMDs) and 2) into two nucleotide-binding domains (NBDs). Both intramolecular domains are coupled to each other and when ATP binding is in progress, conformational changes in the TMDs allow proteasomal degradation products to move across the membrane. TAP recognizes and transports the antigen peptides produced in the cytosol straight into the ER, while tapasin recognizes the kind of peptides that have the ability to form stable complexes with MHC-I. This process is known as peptide proofreading or editing. Peptides selected through proofreading[2] improve MHC-I stability; tapasin also contributes to the editing of immunogenic peptide epitopes. However, only lately it was proven via biochemical, biophysical, and structural studies that a key function in adaptive immunity, the catalytic mechanism of peptide proofreading, is performed by tapasin and TAPBPR (TAP-binding protein-related, a tapasin homologue).[3]
Tapasin
Cresswell and co-workers first discovered tapasin (TAP-associated glycoprotein) as a 48 kDa protein in complexes isolated with TAP1 antibodies from digitonin lysates of human B lymphoblastoid cells.[4] Tapasin binds HC/β2m along with ER chaperones to the peptide transporter.[5] It is located in the ER and its function comprises holding together class I molecules jointly with the chaperone calreticulin and the ERp57 to TAP. Studies of a tapasin-deficient cell line and from mice bearing a disrupted tapasin gene, the short-lived complex of class I molecules.
Tapasin and TAP are very important for the stabilization of the class I molecules and also for the optimization of the peptide presented to cytotoxic T cells.[6] A PLC-independent tapasin homologue protein named TAPBPR[3] was found that has the ability to act as a second MHC-I specific peptide proofreader or editor, but does not possess a transmembrane domain.[7] Tapasin and TAPBPR[3] share similar binding interfaces on MHC-I, as shown with the X-ray structure of TAPBPR with MHC-I (heavy chain and β2 microglobulin). The use of a photo-cleavable high-affinity peptide allowed researchers to form a stable (bound) MHC-I molecules and afterwards to form a stable TAPBPR[3] and MHC-I complex with cleavage by UV light of the photoinduced peptide.
ERp57
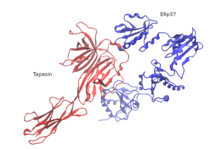
ERp57 is an enzyme of the thiol oxidoreductase family located in the ER.[8] It is attached to substrates in an indirect fashion through association with the molecular chaperone calreticulin of the peptide-loading complex,[9][10] In early stages of generation of MHC-I molecules, ERp57 is associated with free MHC-I heavy chains. As a result, its function is determined by the formation of disulfide bonds in heavy chains, by oxidative folding of the heavy chain, and finally by the fact that ERp57 is loading the peptides onto MHC-I molecules.
MHC-I
Preliminary MHC-I heavy chains form chaperones with the aid of the calnexin-calreticulin complex in the ER. In addition to this, β2-microglobulin (β2m) is attached to the heavy chains of the heterodimers and as a whole they act as receptors for antigenic peptides. When MHC-I chains are empty, they are recruited by calreticulin and form a transient PLC.
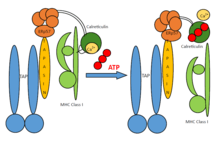
Tapasin regularly plays a role in the stabilization of MHC-I. Only after MHC-I heterodimers are deployed for peptide proofreading or editing, stable pMHC-I (peptide-MHC-I) complexes are released to the cell surface for recognition and destruction of virus-infected or malignantly neoplastic cells. In general, each individual organism owns a collection of six MHC-I molecules (three from each parent). Thus, in autoimmune emergencies, compatible donors are relatives who own a similar collection of MHC-I molecules, apart from those of the recipient.
Calreticulin
Calreticulin – especially its lectin-like domain – interacts with MHC-I. The P domain faces the MHC-I peptide-binding site towards ERp57. This orientation makes it possible for tapasin to attach and secure MHC-I. This translocation of TAP facilitates its opening out into an ER luminal cavity, edged by standard membrane entry points such as those for tapasin and MHC-I. These two entry points facilitate the recruitment of MHC-I with optimal peptide loading and eventual release of MHC-I in T-cell surfaces for recognition.
References
- Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampé R (November 2017). "Structure of the human MHC-I peptide-loading complex". Nature. 551 (7681): 525–528. Bibcode:2017Natur.551..525B. doi:10.1038/nature24627. PMID 29107940.
- Thomas C, Tampé R (2017). "Proofreading of Peptide-MHC Complexes through Dynamic Multivalent Interactions". Frontiers in Immunology. 8: 65. doi:10.3389/fimmu.2017.00065. PMC 5296336. PMID 28228754.
- Thomas C, Tampé R (November 2017). "Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing". Science. 358 (6366): 1060–1064. Bibcode:2017Sci...358.1060T. doi:10.1126/science.aao6001. PMID 29025996.
- Ortmann B, Androlewicz MJ, Cresswell P (April 1994). "MHC class I/beta 2-microglobulin complexes associate with TAP transporters before peptide binding". Nature. 368 (6474): 864–7. Bibcode:1994Natur.368..864O. doi:10.1038/368864a0. PMID 8159247.
- Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P (August 1996). "Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP". Immunity. 5 (2): 103–14. doi:10.1016/S1074-7613(00)80487-2. PMID 8769474.
- Momburg F, Tan P (October 2002). "Tapasin-the keystone of the loading complex optimizing peptide binding by MHC class I molecules in the endoplasmic reticulum". Molecular Immunology. 39 (3–4): 217–33. doi:10.1016/S0161-5890(02)00103-7. PMID 12200052.
- McShan AC, Natarajan K, Kumirov VK, Flores-Solis D, Jiang J, Badstübner M, et al. (August 2018). "Peptide exchange on MHC-I by TAPBPR is driven by a negative allostery release cycle". Nature Chemical Biology. 14 (8): 811–820. doi:10.1038/s41589-018-0096-2. PMC 6202177. PMID 29988068.
- Frickel EM, Frei P, Bouvier M, Stafford WF, Helenius A, Glockshuber R, Ellgaard L (April 2004). "ERp57 is a multifunctional thiol-disulfide oxidoreductase". The Journal of Biological Chemistry. 279 (18): 18277–87. doi:10.1074/jbc.M314089200. PMID 14871896.
- Oliver JD, Roderick HL, Llewellyn DH, High S (August 1999). "ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin". Molecular Biology of the Cell. 10 (8): 2573–82. doi:10.1091/mbc.10.8.2573. PMC 25489. PMID 10436013.
- Zhang Y, Baig E, Williams DB (May 2006). "Functions of ERp57 in the folding and assembly of major histocompatibility complex class I molecules". The Journal of Biological Chemistry. 281 (21): 14622–31. doi:10.1074/jbc.M512073200. PMID 16567808.