Pan-genome
In the fields of molecular biology and genetics, a pan-genome (or supragenome) is the entire set of genes for all strains within a clade.[1][2][3] The pan-genome includes: the core genome containing genes present in all strains within the clade, the accessory genome containing 'dispensable' genes present in a subset of the strains, and strain-specific genes.[1][2][3] The study of the pan-genome is called pangenomics.[1]
Some species have open (or extensive) pan-genomes, while others have closed pan-genomes.[1] For species with a closed pan-genome, very few genes are added per sequenced genome (after sequencing many strains), and the size of the full pan-genome can be theoretically predicted. Species with an open pan-genome have enough genes added per additional sequenced genome that predicting the size of the full pan-genome is impossible.[3] Population size and niche versatility have been suggested as the most influential factors in determining pan-genome size.[1] The pan-genome can be broken down into a "core pan-genome" that contains genes present in all individuals, a "shell pan-genome" that contains genes present in two or more strains, and a "cloud pan-genome" that contains genes only found in a single strain.[2][3][4][5]
Pan-genomes were originally constructed for species of bacteria and archaea, but more recently eukaryotic pan-genomes have been developed, particularly for plant species. Plant studies have shown that pan-genome dynamics are linked to transposable elements.[6][7][8] The significance of the pan-genome arises in an evolutionary context, especially with relevance to metagenomics,[9] but is also used in a broader genomics context.[10]
An open access book reviewing the pan-genome concept and its implications, edited by Tettelin and Medini, was published in the spring of 2020.[11]
History
Etymology
The term ‘pan-genome’ was defined with its current meaning by Tettelin et al. in 2005;[1] it derives 'pan' from the Greek word παν, meaning 'whole' or 'everything', while genome is a commonly used term to describe an organism's complete genetic material. Tettelin et al. applied the term specifically to bacteria, whose pan-genome "includes a core genome containing genes present in all strains and a dispensable genome composed of genes absent from one or more strains and genes that are unique to each strain."[1]
Original concept
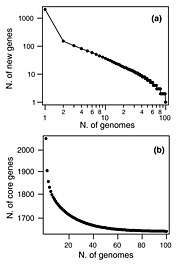
The original pan-genome concept was developed by Tettelin et al.[1] when they analyzed the genomes of eight isolates of Streptococcus agalactiae which could be described as a core genome shared by all isolates, accounting for approximately 80% of any single genome, plus a dispensable genome consisting of partially shared and strain-specific genes. Extrapolation suggested that the gene reservoir in the S. agalactiae pan-genome is vast and that new unique genes would continue to be identified even after sequencing hundreds of genomes.[1]
Data structures
Pangenome graphs are emerging data structures designed to represent pangenomes and to efficiently map reads to them. They have been reviewed by Eizenga et al [13]
Examples
A similar pattern was found in Streptococcus pneumoniae when 44 strains were sequenced (see figure). With each new genome sequenced fewer new genes were discovered. In fact, the predicted number of new genes dropped to zero when the number of genomes exceeds 50 (note, however, that this is not a pattern found in all species). This would mean that S. pneumoniae has a 'closed pan-genome'.[14] The main source of new genes in S. pneumoniae was Streptococcus mitis from which genes were transferred horizontally. The pan-genome size of S. pneumoniae increased logarithmically with the number of strains and linearly with the number of polymorphic sites of the sampled genomes, suggesting that acquired genes accumulate proportionately to the age of clones.[12]
Another example for the latter can be seen in a comparison of the sizes of the core and the pan-genome of Prochlorococcus. The core genome set is logically much smaller than the pan-genome, which is used by different ecotypes of Prochlorococcus.[15] A 2015 study on Prevotella bacteria isolated from humans, compared the gene repertoires of its species derived from different body sites of human. It also reported an open pan- genome showing vast diversity of gene pool.[16]. Open pan-genome has been observed in environmental isolates such as Alcaligenes sp.[17] and Serratia sp. [18], showing a sympatric lifestyle.
Software tools
As interest in pan-genomes increased, there have been a number of software tools developed to help analyze this kind of data. In 2015, a group reviewed the different kinds of analyses and tools a researcher may have available.[19] There are seven kinds of analyses software developed to analyze pangenomes: cluster homologous genes; identify SNPs; plot pangenomic profiles; build phylogenetic relationships of orthologous genes/families of strains/isolates; function-based searching; annotation and/or curation; and visualizations.[19]
The two most cited software tools at the end of 2014[19] were Panseq[20] and the pan-genomes analysis pipeline (PGAP).[21] Other options include BPGA – A Pan-Genome Analysis Pipeline for prokaryotic genomes,[22] GET_HOMOLOGUES ,[23] Roary[24] and PanDelos.[25]
A review focused on plant pan-genomes was published in 2015.[26] Among the first software packages designed for plant pan-genomes were PanTools[27] and GET_HOMOLOGUES-EST.[8][28]
See also
References
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. (September 2005). "Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome"". Proceedings of the National Academy of Sciences of the United States of America. 102 (39): 13950–5. Bibcode:2005PNAS..10213950T. doi:10.1073/pnas.0506758102. PMC 1216834. PMID 16172379.
- Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R (December 2005). "The microbial pan-genome". Current Opinion in Genetics & Development. 15 (6): 589–94. doi:10.1016/j.gde.2005.09.006. PMID 16185861.
- Vernikos G, Medini D, Riley DR, Tettelin H (February 2015). "Ten years of pan-genome analyses". Current Opinion in Microbiology. 23: 148–54. doi:10.1016/j.mib.2014.11.016. PMID 25483351.
- Wolf YI, Makarova KS, Yutin N, Koonin EV (December 2012). "Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer". Biol. Direct. 7: 46. doi:10.1186/1745-6150-7-46. PMC 3534625. PMID 23241446.
- Vernikos, George; Medini, Duccio; Riley, David R; Tettelin, Hervé (2015). "Ten years of pan-genome analyses". Current Opinion in Microbiology. 23: 148–154. doi:10.1016/j.mib.2014.11.016. PMID 25483351.
- Morgante M, De Paoli E, Radovic S (April 2007). "Transposable elements and the plant pan-genomes". Current Opinion in Plant Biology. 10 (2): 149–55. doi:10.1016/j.pbi.2007.02.001. PMID 17300983.
- Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, et al. (December 2017). "Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure". Nature Communications. 8 (1): 2184. Bibcode:2017NatCo...8.2184G. doi:10.1038/s41467-017-02292-8. PMC 5736591. PMID 29259172.
- Contreras-Moreira B, Cantalapiedra CP, García-Pereira MJ, Gordon SP, Vogel JP, Igartua E, et al. (February 2017). "Analysis of Plant Pan-Genomes and Transcriptomes with GET_HOMOLOGUES-EST, a Clustering Solution for Sequences of the Same Species". Frontiers in Plant Science. 8: 184. doi:10.3389/fpls.2017.00184. PMC 5306281. PMID 28261241.
- Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ (May 2009). "Biogeography of the Sulfolobus islandicus pan-genome". Proceedings of the National Academy of Sciences of the United States of America. 106 (21): 8605–10. Bibcode:2009PNAS..106.8605R. doi:10.1073/pnas.0808945106. PMC 2689034. PMID 19435847.
- Reinhardt JA, Baltrus DA, Nishimura MT, Jeck WR, Jones CD, Dangl JL (February 2009). "De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae". Genome Research. 19 (2): 294–305. doi:10.1101/gr.083311.108. PMC 2652211. PMID 19015323.
- Tettelin, Hervé; Medini, Duccio, eds. (2020). The Pangenome (PDF). doi:10.1007/978-3-030-38281-0. ISBN 978-3-030-38280-3. S2CID 217167361.
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, et al. (2010). "Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species". Genome Biology. 11 (10): R107. doi:10.1186/gb-2010-11-10-r107. PMC 3218663. PMID 21034474.
- Eizenga JM, Novak AM, Sibbesen JA, Heumos S, Ghaffaari A, Hickey G, Chang X, Seaman JD, Rounthwaite R, Ebler J, Rautiainen M, Garg S, Paten B, Marschall T, Sirén T, Garrison E (August 2020). "Pangenome Graphs". Annual Review of Genomics and Human Genetics. 21. doi:10.1146/annurev-genom-120219-080406. PMID 32453966.
- Rouli L, Merhej V, Fournier PE, Raoult D (September 2015). "The bacterial pangenome as a new tool for analysing pathogenic bacteria". New Microbes and New Infections. 7: 72–85. doi:10.1016/j.nmni.2015.06.005. PMC 4552756. PMID 26442149.
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, et al. (December 2007). "Patterns and implications of gene gain and loss in the evolution of Prochlorococcus". PLOS Genetics. 3 (12): e231. doi:10.1371/journal.pgen.0030231. PMC 2151091. PMID 18159947.
- Gupta VK, Chaudhari NM, Iskepalli S, Dutta C (March 2015). "Divergences in gene repertoire among the reference Prevotella genomes derived from distinct body sites of human". BMC Genomics. 16 (153): 153. doi:10.1186/s12864-015-1350-6. PMC 4359502. PMID 25887946.
- Basharat Z, Yasmin A, He T, Tong Y (2018). "Genome sequencing and analysis of Alcaligenes faecalis subsp. phenolicus MB207". Scientific Reports. 8 (1): 3616. Bibcode:2018NatSR...8.3616B. doi:10.1038/s41598-018-21919-4. PMC 5827749. PMID 29483539.
- Basharat Z, Yasmin A (2016). "Pan-genome Analysis of the Genus Serratia". arXiv:1610.04160 [q-bio.GN].
- Xiao J, Zhang Z, Wu J, Yu J (February 2015). "A brief review of software tools for pangenomics". Genomics, Proteomics & Bioinformatics. 13 (1): 73–6. doi:10.1016/j.gpb.2015.01.007. PMC 4411478. PMID 25721608.
- Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, et al. (September 2010). "Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions". BMC Bioinformatics. 11 (1): 461. doi:10.1186/1471-2105-11-461. PMC 2949892. PMID 20843356.
- Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J (February 2012). "PGAP: pan-genomes analysis pipeline". Bioinformatics. 28 (3): 416–8. doi:10.1093/bioinformatics/btr655. PMC 3268234. PMID 22130594.
- Chaudhari NM, Gupta VK, Dutta C (April 2016). "BPGA- an ultra-fast pan-genome analysis pipeline". Scientific Reports. 6 (24373): 24373. Bibcode:2016NatSR...624373C. doi:10.1038/srep24373. PMC 4829868. PMID 27071527.
- Contreras-Moreira B, Vinuesa P (December 2013). "GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis". Applied and Environmental Microbiology. 79 (24): 7696–701. doi:10.1128/AEM.02411-13. PMC 3837814. PMID 24096415.
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. (November 2015). "Roary: rapid large-scale prokaryote pan genome analysis". Bioinformatics. 31 (22): 3691–3. doi:10.1093/bioinformatics/btv421. PMC 4817141. PMID 26198102.
- Bonnici V, Giugno R, Manca V (November 2018). "PanDelos: a dictionary-based method for pan-genome content discovery". BMC Bioinformatics. 19 (Suppl 15): 437. doi:10.1186/s12859-018-2417-6. PMC 6266927. PMID 30497358.
- Golicz AA, Batley J, Edwards D (April 2016). "Towards plant pangenomics" (PDF). Plant Biotechnology Journal. 14 (4): 1099–105. doi:10.1111/pbi.12499. PMID 26593040.
- Sheikhizadeh S, Schranz ME, Akdel M, de Ridder D, Smit S (September 2016). "PanTools: Representation, Storage and Exploration of Pan-Genomic Data". Bioinformatics. 32 (17): i487–i493. doi:10.1093/bioinformatics/btw455. PMID 27587666.
- Contreras-Moreira B, Vinuesa P (December 2013). "GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis". Applied and Environmental Microbiology. 79 (24): 7696–701. doi:10.1128/AEM.02411-13. PMC 3837814. PMID 24096415.