PBAD promoter
PBAD (systematically araBp) is a promoter found in bacteria and especially as part of plasmids used in laboratory studies. The promoter is a part of the arabinose operon whose name derives from the genes it regulates transcription of: araB, araA, and araD.[1][2] In E. coli, the PBAD promoter is adjacent to the PC promoter (systematically araCp), which transcribes the araC gene in the opposite direction. araC encodes the AraC protein, which regulates activity of both the PBAD and PC promoters.[3][4][5] The cyclic AMP receptor protein CAP binds between the PBAD and PC promoters, stimulating transcription of both when bound by cAMP.[6]

Regulation of PBAD
Transcription initiation at the PBAD promoter occurs in the presence of high arabinose and low glucose concentrations.[7] Upon arabinose binding to AraC, the N-terminal arm of AraC is released from its DNA binding domain via a “light switch” mechanism.[1][2] This allows AraC to dimerize and bind the I1 and I2 operators.[3] The AraC-arabinose dimer at this site contributes to activation of the PBAD promoter.[2] Additionally, CAP binds to two CAP binding sites upstream of the I1 and I2 operators and helps activate the PBAD promoter.[6] In the presence of both high arabinose and high glucose concentrations however, low cAMP levels prevent CAP from activating the PBAD promoter.[7] It is hypothesized that PBAD promoter activation by CAP and AraC is mediated through contacts between the C-terminal domain of the α-subunit of RNA polymerase and the CAP and AraC proteins.[8]
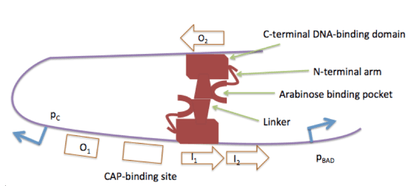
Without arabinose, and regardless of glucose concentration, the PBAD and PC promoters are repressed by AraC.[2][7] The N-terminal arm of AraC interacts with its DNA binding domain, allowing two AraC proteins to bind to the O2 and I1 operator sites.[1] The O2 operator is situated within the araC gene. An AraC dimer also binds to the O1 operator and represses the PC promoter via a negative autoregulatory feedback loop.[2] The two bound AraC proteins dimerize and cause looping of the DNA.[3][4] The looping prevents binding of CAP and RNA Polymerase, which normally activate the transcription of both PBAD and PC.
|
Transcription by PBAD |
High Arabinose |
Low Arabinose |
|
High Glucose |
Repressed |
Repressed |
|
Low Glucose |
Active |
Repressed |
The spacing between the O2 and I1 operator sites is critical. Adding or removing 5 base pairs between the O2 and I1 operator sites abrogates AraC mediated repression of the PBAD promoter.[1] The spacing requirement arises from the double helix nature of DNA, in which a complete turn of the helix is about 10.5 nucleotides. Therefore, adding or removing 5 base pairs between the O2 and I1 operator sites rotates the helix roughly 180 degrees. This reverses the direction that the O2 operator faces when the DNA is looped and prevents dimerization of the O2 bound AraC with the bound I1 araC.[1][2]
The PBAD promoter on expression plasmids

The PBAD promoter allows for tight regulation and control of a target gene in vivo.[7] As explained above, PBAD is regulated by the addition and absence of arabinose. As tested, the promoter can be further repressed with reduced levels of cAMP through the addition of glucose.[7] Plasmid vectors have been constructed and tested with a selectable marker (CmR in this case), origin of replication, araC and operons, multiple cloning site and PBAD promoter. Studies show that vectors are highly expressed and can be used, in combination with chromosomal null alleles, to study loss of function of essential genes.[7]
References
- Schleif R. AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev (2010) 1–18.
- Schleif R. AraC protein: a love-hate relationship. Bioessays 2003, 25:274-282.
- Reed WL, Schleif RF. Hemiplegic Mutations in AraC Protein. J. Mol. Biol. (1999) 294, 417-425.
- Lobell RB, Schleif RF. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528-32.
- Soisson SM, MacDougall-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science. 1997 Apr 18;276(5311):421-5.
- Dunn T.M., Schleif R. Deletion Analysis of the Escherichia coli ara PC and PBAD Promoters. J.Mol. Biol. (1984) 180, 201-204.
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. Jul 1995; 177(14): 4121–4130.
- Johnson CM, Schleif RF. Cooperative Action of the Catabolite Activator Protein and AraC In Vitro at the araFGH Promoter. J Bacteriol. Apr 2000; 182(7).