P-TEFb
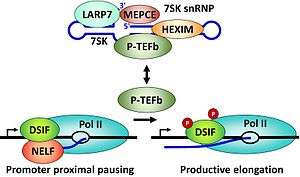
The positive transcription elongation factor, P-TEFb, is a multiprotein complex that plays an essential role in the regulation of transcription by RNA polymerase II (Pol II) in eukaryotes.[1] Immediately following initiation Pol II becomes trapped in promoter proximal paused positions on the majority of human genes (Figure 1).[2][3] P-TEFb is a cyclin dependent kinase that can phosphorylate the DRB sensitivity inducing factor (DSIF)[4] and negative elongation factor (NELF),[5] as well as the carboxyl terminal domain of the large subunit of Pol II[6] and this causes the transition into productive elongation leading to the synthesis of mRNAs. P-TEFb is regulated in part by a reversible association with the 7SK snRNP.[7] Treatment of cells with the P-TEFb inhibitors DRB or flavopidirol leads to loss of mRNA production and ultimately cell death.[6][8]
Discovery, Composition and Structure
P-TEFb was identified and purified as a factor needed for the generation of long run-off transcripts using an in vitro transcription system derived from Drosophila cells.[9] It is a cyclin dependent kinase containing the catalytic subunit, Cdk9, and a regulatory subunit, cyclin T in Drosophila.[10] In humans there are multiple forms of P-TEFb which contain Cdk9 and one of several cyclin subunits, cyclin T1, T2, and K.[11][12] P-TEFb associates with other factors including the bromodomain protein BRD4,[13] and is found associated with a large complex of proteins called the super elongation complex.[14][15] Importantly, for the AIDS virus, HIV, P-TEFb is targeted by the HIV Tat protein[16] which bypasses normal cellular P-TEFb control and directly brings P-TEFb to the promoter proximal paused polymerase in the HIV genome.[17][18]
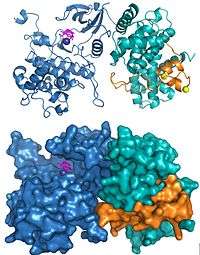
The structures of human P-TEFb containing Cdk9 and cyclin T1 and the HIV Tat•P-TEFb complex have been solved using X-ray crystallography. The first structure solved demonstrated that the two subunits were arranged as has been found in other cyclin dependent kinases.[19] Three amino acid substitutions were inadvertently introduced in the subunits used for the original structure and a subsequent structure determination using the correct sequences demonstrated the same overall structure except for a few significant changes around the active site.[20] The structure of HIV Tat bound to P-TEFb demonstrated that the viral protein forms extensive contacts with the cyclin T1 subunit (Figure 2).[20]
Regulation of P-TEFb
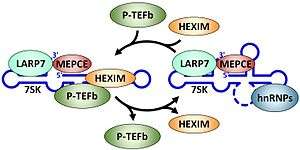
Because of its central role in controlling eukaryotic gene expression, P-TEFb is subject to stringent regulation at the level of transcription of the genes encoding the subunits, translation of the subunit mRNAs, turnover of the subunits, and also by an unusual mechanism involving the 7SK snRNP.[7] As shown in Figure 3 P-TEFb is held in the 7SK snRNP by the double stranded RNA binding protein HEXIM (HEXIM1 or HEXIM2 in humans). HEXIM bound to 7SK RNA or any double stranded RNA binds to P-TEFb and inhibits the kinase activity.[21][22] Two other proteins are always found associated with 7SK RNA. The methyl phosphase capping enzyme MEPCE puts a methyl group on the gamma phosphate of the first nucleotide of the 7SK RNA[23] and the La related protein LARP7 binds to the 3' end of 7SK.[24][25] When P-TEFb is extracted from the 7SK snRNP, 7SK RNA undergoes a conformation change, HEXIM is ejected and hnRNPs take the place of the factors removed.[7] The re-sequestration of P-TEFb requires another rearrangement of the RNA, binding of HEXIM and then P-TEFb. In rapidly growing cells the 7SK snRNP is the predominate form of P-TEFb. For review.[26]
References
- Zhou Q, Li T, Price DH. RNA Polymerase II Elongation Control. Annu Rev Biochem 2012.
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell 2010; 141:432-45.
- Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell 2012; 45:38-50.
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 1998; 12:343-56.
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 1999; 97:41-51.
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 1996; 271:27176-83.
- Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA 2012; 3:92-103.
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 2001; 276:31793-9.
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 1995; 270:12335-8.
- Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J Biol Chem 1998; 273:13855-60.
- Fu TJ, Peng J, Lee G, Price DH, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem 1999; 274:34527-30.
- Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 1998; 12:755-62.
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005; 19:535-45.
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 2011; 25:661-72.
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 2010; 38:428-38.
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 1987; 330:489-93.
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, et al. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev 1997; 11:2622-32.
- Garber ME, Wei P, Jones KA. HIV-1 Tat interacts with cyclin T1 to direct the P-TEFb CTD kinase complex to TAR RNA. Cold Spring Harbor symposia on quantitative biology 1998; 63:371-80.
- Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J 2008; 27:1907-18.
- Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 2010; 465:747-51.
- Li Q, Cooper JJ, Altwerger GH, Feldkamp MD, Shea MA, Price DH. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res 2007; 35:2503-12.
- Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J 2004; 23:2608-19.
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 2007; 27:262-74.
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res 2008; 36:2219-29.
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 2008; 29:588-99.
- Quaresma, AJ; Bugai A; Barboric M. (2016). "Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb". Nucleic Acids Research. 44 (8): 7527–7539. doi:10.1093/nar/gkw585. PMC 5027500. PMID 27369380.