Methylglyoxal pathway
The methylglyoxal pathway is an offshoot of glycolysis found in some prokaryotes, which converts glucose into methylglyoxal and then into pyruvate. However unlike glycolysis the methylglyoxal pathway does not produce adenosine triphosphate, ATP. The pathway is named after the substrate methylglyoxal which has three carbons and two carbonyl groups located on the 1st carbon and one on the 2nd carbon. Methylglyoxal is, however, a reactive aldehyde that is very toxic to cells, it can inhibit growth in E. coli at milimolar concentrations. The excessive intake of glucose by a cell is the most important process for the activation of the methylglyoxal pathway.
The Methylglyoxal pathway
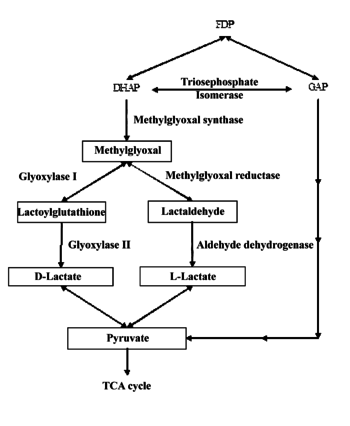
The methylglyoxal pathway is activated by the increased intercellular uptake of carbon containing molecules such as glucose, glucose-6-phosphate, lactate, or glycerol. Methylglyoxal is formed from dihydroxyacetone phosphate (DHAP) by the enzyme methylglyoxal synthase, giving off a phosphate group.
Methylglyoxal is then converted into two different products, either D-lactate, and L-lactate. Methylglyoxal reductase and aldehyde dehydrogenase convert methylglyoxal into lactaldehyde and, eventually, L-lactate. If methylglyoxal enters the glyoxylase pathway, it is converted into lactoylguatathione and eventually D-lactate. Both D-lactate, and L-lactate are then converted into pyruvate. The pyruvate that is created most often goes on to enter the Krebs cycle (Weber 711-13).
Enzymes and regulation
The potentially hazardous effects of methylglyoxal require regulation of the reactions with this substrate. Synthesis of methylglyoxal is regulated by levels of DHAP and phosphate concentrations. High concentrations of DHAP encourage methylglyoxal synthase to produce methylglyoxal, while high phosphate concentrations inhibit the enzyme, and therefore the production of more methylglyoxal. The enzyme triose phosphate isomerase affects the levels of DHAP by converting glyceraldehyde 3-phosphate (GAP) into DHAP. The usual pathway converting GAP to pyruvate starts with the enzyme glyceraldehyde 3-phosphate dehydrogenase (Weber 711-13). Low phosphate levels inhibit GAP dehydrogenase; GAP is instead converted into DHAP by triosephosphate isomerase. Again, increased levels of DHAP activate methylglyoxal synthase and methylglyoxal production (Weber 711-13).
The oscillation of Methylglyoxal concentration in feast concentrations
Jan Weber, Anke Kayser, and Ursula Rinas, performed an experiment to test what happened to the methylglyoxal pathway when E. coli was in the presence of a constantly high concentration of glucose. The concentration of methylglyoxal increased until it reached 20 μmol. Methylglyoxal concentration then began to decrease, once it reached this level. The decrease in the concentration of methylglyoxal was connected to the drop in respiratory activity. When respiration activity increased the concentration of methylglyoxal increased again, until it reached the 20 μmol concentration (Weber 714-15).
Why does the Methylglyoxal pathway exist?
This pathway does not produce any ATP, this pathway does not replace glycolysis, it runs simultaneously to glycolysis and is only initiated with an increased concentration of sugar phosphates. One believed purpose of the methylglyoxal pathway is to help release the stress of elevated sugar phosphate concentration. Also when methylglyoxal is formed from DHAP, an inorganic phosphate is given off which can be used to replenish a low concentration of needed inorganic phosphate. The methylglyoxal pathway is a rather dangerous tactic, both because less energy is produced and a toxic compound, methylglyoxal is formed. (Weber 715).
References
Weber, Jan, Anke Kayser, and Ursula Rinas. Metabolic Flux Analysis of Escherichia Coli In. Vers. 151: 707-716. 6 Dec. 2004. Microbiology. 10 Apr. 2007 <http://mic.sgmjournals.org/cgi/reprint/151/3/707>.
Saadat, D., Harrison, D.H.T. "Methylglyoxal Synthase From Escherichia Coli." RCSB Protein Data Base. 24 Apr. 2007. RCSB Protein Data Base. 25 Apr. 2007 <http://www.pdb.org/pdb/explore.do?structureId=1B93>.
"Methylglyoxal Synthase From Escherichia Coli." RCSB Protein Data Base. 24 Apr. 2007. RCSB Protein Data Base. 25 Apr. 2007 <http://www.pdb.org/pdb/explore.do?structureId=1B93>.
Yun, M., C.-G. Park, J.-Y Kim, and H.-W. Park. "Structural Anayysis of Glyeraldehyde 3-Phosphate Dehydrogenase from Escherichia coli: Direct Evidence for Substrate Binding and Cofactor-Induced Confromational Changes. RCSB Protein Data Base. 24 Apr. 2007. RCSB Protein Data Base. 30 Apr. 2007 <http://www.pdb.org/pdb/explore.do?structureId=1DC4>.