Magnetic resonance elastography
Magnetic resonance elastography (MRE) is a non-invasive medical imaging technique that measures the stiffness of soft tissues by generating shear waves in tissue, imaging their propagation using MRI, and processing the images to generate a stiffness map (elastogram).[1] It is one of the most commonly used elastography techniques.[2]
| Magnetic resonance elastography | |
|---|---|
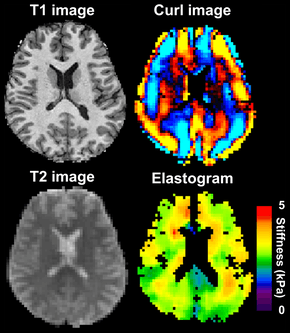 Magnetic resonance elastography of the brain. A T1 weighted anatomical image is shown in the top-left, and the corresponding T2 weighted image from the MRE data is shown in the bottom-left. The wave image used to make the elastogram is shown in the top-right, and the resulting elastogram is in the bottom-right. | |
| Purpose | measures the mechanical properties of soft tissues |
MRE was first described by Muthupillai et al. in 1995.[3] Because diseased tissues are often stiffer than the surrounding normal tissue, MRE has been applied to visualize a variety of disease processes which affect tissue stiffness in the liver, breast, brain, heart, and skeletal muscle.[1][4] For example, breast tumors are much harder than healthy fibroglandular tissue.[5] MRE is similar to palpation; however, whereas palpation is a qualitative technique performed by physicians, MRE is a quantitative technique performed with a radiologist.[1]
Mechanics of Soft Tissue
MRE quantitatively determines the stiffness of biological tissues by measuring its mechanical response to an external stress.[4] Specifically, MRE calculates the shear modulus of a tissue from its shear-wave displacement measurements.[3] The elastic modulus quantifies the stiffness of a material, or how well it resists elastic deformation as a force is applied. For elastic materials, strain is directly proportional to stress within an elastic region. The elastic modulus is seen as the proportionality constant between stress and strain within this region. Unlike purely elastic materials, biological tissues are viscoelastic, meaning that it has characteristics of both elastic solids and viscous liquids. Their mechanical responses depend on the magnitude of the applied stress as well as the strain rate. The stress-strain curve for a viscoelastic material exhibits hysteresis. The area of the hysteresis loop represents the amount of energy lost as heat when a viscoelastic material undergoes an applied stress and is distorted. For these materials, the elastic modulus is complex and can be separated into two components: a storage modulus and a loss modulus. The storage modulus expresses the contribution from elastic solid behavior while the loss modulus expresses the contribution from viscous liquid behavior. Conversely, elastic materials exhibit a pure solid response. When a force is applied, these materials elastically store and release energy, which does not result in energy loss in the form of heat.[6]
Yet, MRE and other elastography imaging techniques typically utilize a mechanical parameter estimation that assumes biological tissues to be linearly elastic and isotropic for simplicity purposes.[1] The effective shear modulus can be expressed with the following equation:
where is the elastic modulus of the material and is the Poisson’s ratio.
The Poisson’s ratio for soft tissues is approximated to equal 0.5, resulting in the ratio between the elastic modulus and shear modulus to equal 3.[7] This relationship can be used to estimate the stiffness of biological tissues based on the calculated shear modulus from shear-wave propagation measurements. A driver system produces and transmits acoustic waves set at a specific frequency (50–500 Hz) to the tissue sample. At these frequencies, the velocity of shear waves can be about 1–10 m/s.[8][9] The effective shear modulus can be calculated from the shear wave velocity with the following:[10]
where is the tissue density and is the shear wave velocity.
Recent studies have been focused on incorporating mechanical parameter estimations into post-processing inverse algorithms that account for the complex viscoelastic behavior of soft tissues. Creating new parameters could potentially increase the specificity of MRE measurements and diagnostic testing.[11][12]
Applications
Liver
Liver fibrosis is a common result of many chronic liver diseases; progressive fibrosis can lead to cirrhosis. MRE of the liver provides quantitative maps of tissue stiffness over large regions of the liver. This non-invasive technique is able to detect increased stiffness of the liver parenchyma, which is a direct consequence of liver fibrosis. It helps to stage liver fibrosis or diagnose mild fibrosis with reasonable accuracy.[13][14][12][15]
Brain
MRE of the brain was first presented in the early 2000s.[16][17] Elastogram measures have been correlated with memory tasks,[18] fitness measures,[19] and progression of various neurodegenerative conditions. For example, regional and global decreases in brain viscoelasticity have been observed in Alzheimer’s disease[20][21] and multiple sclerosis.[22][23] It has been found that as the brain ages, it loses its viscoelastic integrity due to degeneration of neurons and oligodendrocytes.[24][25] A recent study looked into both the isotropic and anisotropic stiffness in brain and found a correlation between the two and with age, particularly in gray matter.[26]
MRE may also have applications for understanding the adolescent brain. Recently, it was found that adolescents have regional differences in brain viscoelasticity relative to adults.[27][28]
MRE has also been applied to functional neuroimaging. Whereas functional magnetic resonance imaging (fMRI) infers brain activity by detecting relatively slow changes in blood flow, functional MRE is capable of detecting neuromechanical changes in the brain related to neuronal activity occurring on the 100-millisecond scale.[29]
References
- Mariappan YK, Glaser KJ, Ehman RL (July 2010). "Magnetic resonance elastography: a review". Clinical Anatomy. 23 (5): 497–511. doi:10.1002/ca.21006. PMC 3066083. PMID 20544947.
- Chen J, Yin M, Glaser KJ, Talwalkar JA, Ehman RL (April 2013). "MR Elastography of Liver Disease: State of the Art". Applied Radiology. 42 (4): 5–12. PMC 4564016. PMID 26366024.
- Muthupillai R, Lomas DJ, Rossman PJ, Greenleaf JF, Manduca A, Ehman RL (September 1995). "Magnetic resonance elastography by direct visualization of propagating acoustic strain waves". Science. 269 (5232): 1854–7. doi:10.1126/science.7569924. PMID 7569924.
- Glaser KJ, Manduca A, Ehman RL (October 2012). "Review of MR elastography applications and recent developments". Journal of Magnetic Resonance Imaging. 36 (4): 757–74. doi:10.1002/jmri.23597. PMC 3462370. PMID 22987755.
- Pepin KM, Ehman RL, McGee KP (November 2015). "Magnetic resonance elastography (MRE) in cancer: Technique, analysis, and applications". Progress in Nuclear Magnetic Resonance Spectroscopy. 90-91: 32–48. doi:10.1016/j.pnmrs.2015.06.001. PMC 4660259. PMID 26592944.
- Wineman A (2009). "Nonlinear Viscoelastic Solids—A Review". Mathematics and Mechanics of Solids. 14 (3): 300–366. doi:10.1177/1081286509103660. ISSN 1081-2865.
- Low G, Kruse SA, Lomas DJ (January 2016). "General review of magnetic resonance elastography". World Journal of Radiology. 8 (1): 59–72. doi:10.4329/wjr.v8.i1.59. PMC 4731349. PMID 26834944.
- Sarvazyan AP, Skovoroda AR, Emelianov SY, Fowlkes JB, Pipe JG, Adler RS, et al. (1995). "Biophysical Bases of Elasticity Imaging". Acoustical Imaging. Springer US: 223–240. doi:10.1007/978-1-4615-1943-0_23. ISBN 978-1-4613-5797-1.
- Cameron J (1991). "Physical Properties of Tissue. A Comprehensive Reference Book, edited by Francis A. Duck". Medical Physics. 18 (4): 834–834. doi:10.1118/1.596734.
- Wells PN, Liang HD (November 2011). "Medical ultrasound: imaging of soft tissue strain and elasticity". Journal of the Royal Society, Interface. 8 (64): 1521–49. doi:10.1016/S1361-8415(00)00039-6. PMID 21680780.
- Sinkus R, Tanter M, Catheline S, Lorenzen J, Kuhl C, Sondermann E, Fink M (February 2005). "Imaging anisotropic and viscous properties of breast tissue by magnetic resonance-elastography". Magnetic Resonance in Medicine. 53 (2): 372–87. doi:10.1002/mrm.20355. PMID 15678538.
- Asbach P, Klatt D, Schlosser B, Biermer M, Muche M, Rieger A, et al. (October 2010). "Viscoelasticity-based staging of hepatic fibrosis with multifrequency MR elastography". Radiology. 257 (1): 80–6. doi:10.1148/radiol.10092489. PMID 20679447.
- Yin M, Talwalkar JA, Glaser KJ, Manduca A, Grimm RC, Rossman PJ, et al. (October 2007). "Assessment of hepatic fibrosis with magnetic resonance elastography". Clinical Gastroenterology and Hepatology. 5 (10): 1207–1213.e2. doi:10.1016/j.cgh.2007.06.012. PMC 2276978. PMID 17916548.
- Huwart L, Sempoux C, Vicaut E, Salameh N, Annet L, Danse E, et al. (July 2008). "Magnetic resonance elastography for the noninvasive staging of liver fibrosis". Gastroenterology. 135 (1): 32–40. doi:10.1053/j.gastro.2008.03.076. PMID 18471441.
- Venkatesh SK, Yin M, Ehman RL (March 2013). "Magnetic resonance elastography of liver: technique, analysis, and clinical applications". Journal of Magnetic Resonance Imaging. 37 (3): 544–55. doi:10.1002/jmri.23731. PMC 3579218. PMID 23423795.
- Van Houten EE, Paulsen KD, Miga MI, Kennedy FE, Weaver JB (October 1999). "An overlapping subzone technique for MR-based elastic property reconstruction". Magnetic Resonance in Medicine. 42 (4): 779–86. doi:10.1002/(SICI)1522-2594(199910)42:4<779::AID-MRM21>3.0.CO;2-Z. PMID 10502768.
- Van Houten EE, Miga MI, Weaver JB, Kennedy FE, Paulsen KD (May 2001). "Three-dimensional subzone-based reconstruction algorithm for MR elastography". Magnetic Resonance in Medicine. 45 (5): 827–37. doi:10.1002/mrm.1111. PMID 11323809.
- Schwarb H, Johnson CL, McGarry MD, Cohen NJ (May 2016). "Medial temporal lobe viscoelasticity and relational memory performance". NeuroImage. 132: 534–541. doi:10.1016/j.neuroimage.2016.02.059. PMC 4970644. PMID 26931816.
- Schwarb H, Johnson CL, Daugherty AM, Hillman CH, Kramer AF, Cohen NJ, Barbey AK (June 2017). "Aerobic fitness, hippocampal viscoelasticity, and relational memory performance". NeuroImage. 153: 179–188. doi:10.1016/j.neuroimage.2017.03.061. PMC 5637732. PMID 28366763.
- Murphy MC, Huston J, Jack CR, Glaser KJ, Manduca A, Felmlee JP, Ehman RL (September 2011). "Decreased brain stiffness in Alzheimer's disease determined by magnetic resonance elastography". Journal of Magnetic Resonance Imaging. 34 (3): 494–8. doi:10.1002/jmri.22707. PMC 3217096. PMID 21751286.
- Murphy MC, Jones DT, Jack CR, Glaser KJ, Senjem ML, Manduca A, et al. (2016). "Regional brain stiffness changes across the Alzheimer's disease spectrum". NeuroImage. Clinical. 10: 283–90. doi:10.1016/j.nicl.2015.12.007. PMC 4724025. PMID 26900568.
- Streitberger KJ, Sack I, Krefting D, Pfüller C, Braun J, Paul F, Wuerfel J (2012). "Brain viscoelasticity alteration in chronic-progressive multiple sclerosis". PLOS One. 7 (1): e29888. doi:10.1371/journal.pone.0029888. PMC 3262797. PMID 22276134.
- Sandroff BM, Johnson CL, Motl RW (January 2017). "Exercise training effects on memory and hippocampal viscoelasticity in multiple sclerosis: a novel application of magnetic resonance elastography". Neuroradiology. 59 (1): 61–67. doi:10.1007/s00234-016-1767-x. PMID 27889837.
- Sack I, Beierbach B, Wuerfel J, Klatt D, Hamhaber U, Papazoglou S, et al. (July 2009). "The impact of aging and gender on brain viscoelasticity". NeuroImage. 46 (3): 652–7. doi:10.1016/j.neuroimage.2009.02.040. PMID 19281851.
- Sack I, Streitberger KJ, Krefting D, Paul F, Braun J (2011). "The influence of physiological aging and atrophy on brain viscoelastic properties in humans". PLOS One. 6 (9): e23451. doi:10.1371/journal.pone.0023451. PMC 3171401. PMID 21931599.
- Kalra P, Raterman B, Mo X, Kolipaka A (August 2019). "Magnetic resonance elastography of brain: Comparison between anisotropic and isotropic stiffness and its correlation to age". Magnetic Resonance in Medicine. 82 (2): 671–679. doi:10.10002/mrm.27757. PMC 6510588. PMID 30957304.
- Johnson CL, Telzer EH (October 2018). "Magnetic resonance elastography for examining developmental changes in the mechanical properties of the brain". Developmental Cognitive Neuroscience. 33: 176–181. doi:10.1016/j.dcn.2017.08.010. PMC 5832528. PMID 29239832.
- McIlvain G, Schwarb H, Cohen NJ, Telzer EH, Johnson CL (November 2018). "Mechanical properties of the in vivo adolescent human brain". Developmental Cognitive Neuroscience. 34: 27–33. doi:10.1016/j.dcn.2018.06.001. PMC 6289278. PMID 29906788.
- Bridger H (17 April 2019). "Seeing brain activity in 'almost real time'". Harvard Gazette. Retrieved 2019-04-20.
| Wikimedia Commons has media related to Magnetic resonance elastography. |