MED26
Mediator of RNA polymerase II transcription subunit 26 is an enzyme that in humans is encoded by the MED26 gene.[5][6] It forms part of the Mediator complex.
| Med26 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
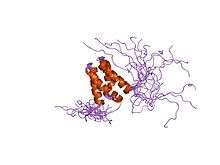 solution structure of the n-terminal domain i of mouse transcription elongation factor s-ii protein 3 | |||||||||
| Identifiers | |||||||||
| Symbol | Med26 N-terminal domain | ||||||||
| Pfam | PF08711 | ||||||||
| InterPro | IPR017923 | ||||||||
| |||||||||
| Mediator subunit 26 Middle domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Med26_M | ||||||||
| Pfam | PF15694 | ||||||||
| |||||||||
| Mediator subunit 26 C-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Med26_C | ||||||||
| Pfam | PF15693 | ||||||||
| |||||||||
The activation of gene transcription is a multistep process that is triggered by factors that recognize transcriptional enhancer sites in DNA. These factors work with co-activators to direct transcriptional initiation by the RNA polymerase II apparatus. The protein encoded by this gene is a subunit of the CRSP (cofactor required for SP1 activation) complex, which, along with TFIID, is required for efficient activation by SP1. This protein is also a component of other multisubunit complexes e.g. thyroid hormone receptor-(TR-) associated proteins which interact with TR and facilitate TR function on DNA templates in conjunction with initiation factors and cofactors.[6]
Activity
MED26 is a transcription elongation factor that increases the overall transcription rate of RNA polymerase II by reactivating transcription elongation complexes that have arrested transcription. It does this through recruiting ELL/EAF- and P-TEFb- containing complexes to promoters via a direct interaction with the N-terminal domain (NTD). The MED26 NTD also binds TFIID, and TFIID and elongation complexes interact with MED26 through overlapping binding sites. [7] MED26 NTD may function as a molecular switch contributing to the transition of Pol II into productive elongation.
The three structural domains of TFIIS are conserved from yeast to human. The 80 or so N-terminal residues form a protein interaction domain containing a conserved motif, which has been called the LW motif because of the invariant leucine and tryptophan residues it contains. Although the N-terminal domain is not needed for transcriptional activity, a similar sequence has been identified in other transcription factors and proteins that are predominantly nuclear localized,:[8][9]
- MED26 (also known as CRSP70 and ARC70), a subunit of the Mediator complex, which is required for the activity of the enhancer-binding protein Sp1.
- Elongin A, a subunit of a transcription elongation factor previously known as SIII. It increases the rate of transcription by suppressing transient pausing of the elongation complex.
- PPP1R10, a nuclear regulatory subunit of protein phosphatase 1 that was previously known as p99, FB19 or PNUTS.
- PIBP, a small hypothetical protein that could be a phosphoinositide binding protein.
- IWS1, which is thought to function in both transcription initiation and elongation. The TFIIS N-terminal domain is a compact four-helix bundle. The hydrophobic core residues of helices 2, 3, and 4 are well conserved among TFIIS domains, although helix 1 is less conserved.[9]
Interactions
MED26 has been shown to interact with MED8,[10] Cyclin-dependent kinase 8,[10] POLR2A,[10] MED12[10] and MED28.[10] It also acts synergistically to mediate the interaction between REST (a Kruppel-type zinc finger transcription factor that binds to a 21-bp RE1 silencing element present in over 900 human genes) and Mediator.[11]
References
- GRCh38: Ensembl release 89: ENSG00000105085 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000045248 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Ryu S, Zhou S, Ladurner AG, Tjian R (Feb 1999). "The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1". Nature. 397 (6718): 446–50. doi:10.1038/17141. PMID 9989412.
- "Entrez Gene: CRSP7 cofactor required for Sp1 transcriptional activation, subunit 7, 70kDa".
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, et al. (2011). "Human mediator subunit MED26 functions as a docking site for transcription elongation factors". Cell. 146 (1): 92–104. doi:10.1016/j.cell.2011.06.005. PMC 3145325. PMID 21729782.
- Booth V, Koth CM, Edwards AM, Arrowsmith CH (October 2000). "Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70". J. Biol. Chem. 275 (40): 31266–8. doi:10.1074/jbc.M002595200. PMID 10811649.
- Ling Y, Smith AJ, Morgan GT (2006). "A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS". Nucleic Acids Res. 34 (8): 2219–29. doi:10.1093/nar/gkl239. PMC 1450333. PMID 16648364.
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC (June 2004). "A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology". Mol. Cell. 14 (5): 685–91. doi:10.1016/j.molcel.2004.05.006. PMID 15175163.
- Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Boyer TG (January 2009). "MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression". J. Biol. Chem. 284 (5): 2648–56. doi:10.1074/jbc.M806514200. PMC 2631966. PMID 19049968.
Further reading
- Näär AM, Beaurang PA, Zhou S, et al. (1999). "Composite co-activator ARC mediates chromatin-directed transcriptional activation". Nature. 398 (6730): 828–32. doi:10.1038/19789. PMID 10235267.
- Booth V, Koth CM, Edwards AM, Arrowsmith CH (2000). "Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70". J. Biol. Chem. 275 (40): 31266–8. doi:10.1074/jbc.M002595200. PMID 10811649.
- Näär AM, Taatjes DJ, Zhai W, et al. (2002). "Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation". Genes Dev. 16 (11): 1339–44. doi:10.1101/gad.987602. PMC 186316. PMID 12050112.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Sato S, Tomomori-Sato C, Banks CA, et al. (2004). "A mammalian homolog of Drosophila melanogaster transcriptional coactivator intersex is a subunit of the mammalian Mediator complex". J. Biol. Chem. 278 (50): 49671–4. doi:10.1074/jbc.C300444200. PMID 14576168.
- Tomomori-Sato C, Sato S, Parmely TJ, et al. (2004). "A mammalian mediator subunit that shares properties with Saccharomyces cerevisiae mediator subunit Cse2". J. Biol. Chem. 279 (7): 5846–51. doi:10.1074/jbc.M312523200. PMID 14638676.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Mo X, Kowenz-Leutz E, Xu H, Leutz A (2004). "Ras induces mediator complex exchange on C/EBP beta". Mol. Cell. 13 (2): 241–50. doi:10.1016/S1097-2765(03)00521-5. PMID 14759369.
- Sato S, Tomomori-Sato C, Parmely TJ, et al. (2004). "A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology". Mol. Cell. 14 (5): 685–91. doi:10.1016/j.molcel.2004.05.006. PMID 15175163.
- Zhang X, Krutchinsky A, Fukuda A, et al. (2005). "MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription". Mol. Cell. 19 (1): 89–100. doi:10.1016/j.molcel.2005.05.015. PMID 15989967.
- Olsen JV, Blagoev B, Gnad F, et al. (2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983.