LRRN3
Leucine-rich repeat neuronal protein 3, also known as neuronal leucine-rich repeat protein 3 (NLRR-3), is a protein that in humans is encoded by the LRRN3 gene.[5][6]
| LRRN3 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | LRRN3, FIGLER5, NLRR-3, NLRR3, leucine rich repeat neuronal 3 | ||||||||||||||||||||||||
| External IDs | MGI: 106036 HomoloGene: 36315 GeneCards: LRRN3 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 7: 111.09 – 111.13 Mb | Chr 12: 41.45 – 41.49 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Gene
The LRRN3 is located on human chromosome 7, at 7q31.1.[7] It contains 6 distinct gt-ag introns, and transcription produces five different mRNAs that appear to differ by truncation of the 3' end. There are only three main transcript variants that actually encode for the LRRN3 protein, with the longest transcript variant being 3744 base pairs in length. All three of these transcript variants have differing lengths and number of exons, but they all have the exon that includes the entire coding sequence for the LRRN3 protein.[8] There are also two paralogs to the LRRN3 gene. These include the LRRN1 and LRRN2 genes, both of which have the same leucine-rich repeats that are characteristic to this family of genes.[5]
Protein
The LRRN3 protein is 708 amino acids in length. The molecular weight of this protein is 79,424 Daltons, with an isoelectric point of 8.02.[9] It is known to be a single-pass type I membrane protein because it spans the membrane once, with its N-terminus on the extracellular side of the membrane, and its signal sequence is removed.[7]
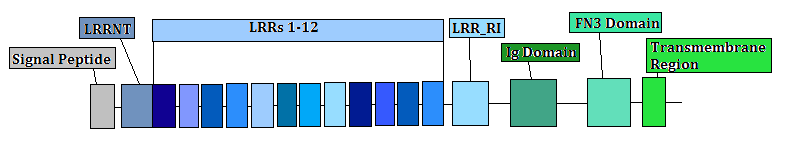
The LRRN3 protein contains 12 leucine-rich repeats, along with an LRRNT and an LRR_RI domain. Leucine-rich repeats are unusually rich in the hydrophobic amino acid leucine. They are common in protein-protein interaction motifs and are typically 20-29 amino acids in length. All major classes of LRRs are known to have a curved horseshoe structure with a parallel beta sheet on the concave side and mostly helical elements on the convex side. The LRRN3 protein also has an immunoglobulin domain, a fibronectin type III domain, and a transmembrane region toward the end of the protein.[10] The composition of this protein is shown below.
Gene Conservation

The LRRN3 gene has been shown to be extremely highly conserved. There are 21 orthologs and homologs for this gene going back to zebra fish. This gene has not been found in any invertebrates or plant species. A multiple sequence alignment has shown this very high conservation of the LRRN3 gene among many different species. All 12 of the leucine-rich repeats, along with the LRRNT and the LRR_RI regions, are highly conserved in all vertebrates for which orthologs of this protein have been obtained. The Ig and FN3 domains also show high conservation in all of the orthologous sequences for mammals and birds, but are not as highly conserved for the rest of the homologous sequences. The high conservation of the leucine-rich repeats, the Ig domain, and the FN3 domain show that these regions must be of importance to the functionality of the LRRN3 gene.[11]
Gene Expression

Gene Expression data has shown that the LRRN3 gene is expressed at very high levels in humans, about 2.3 times the average gene.[8] It is most highly expressed in brain, heart, and testes tissues. It is also slightly expressed in kidney, muscle, pharynx, placental, and thymus tissue. The highest expression of the LRRN3 gene for the developmental stages is the fetal stage, but it is also expressed in the infant, juvenile, and adult stages, as can be seen in the EST profile.[12]
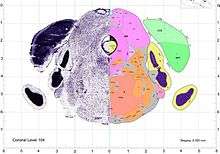
The expression data for the LRRN3 gene shows that it has its highest expression in the brain. Overall, the LRRN3 gene seems to have high to moderate expression throughout the majority of brain tissue. The highest expression being found in the cerebral cortex, the hippocampal formation, the cerebellar cortex, and the paraflocculus. All of these regions play a role in some key cognitive function, involving the processing of language and sensory stimuli, memory, and oculomotor behavior.[13] The high expression of the LRRN3 gene in these regions of the brain could show that the LRRN3 gene has some importance in these cognitive function.
Structure
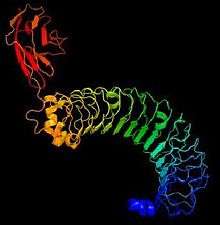
The characteristic shape of leucine-rich repeat proteins is an arc or horseshoe shape. This horseshoe shape of the protein is created by a parallel beta sheet on the concave side and mostly helical elements on the convex side. Eleven residue segments of the LRRs, corresponding to the beta-strand and adjacent loop regions are conserved in LRR proteins, whereas the remaining parts of the repeats may be different. The concave face and the adjacent loops are the most common protein interaction surfaces on LRR proteins. 3D structures of some LRR protein-ligand complexes show that the concave surfact of the LRR domain is ideal for interaction with alpha-helix, thus supporting the conclusions that the elongated and curved LRR structure provides a framework for achieving diverse protein-protein interactions.[10]
Comparison of the LRRN3 protein with the chain A of the crystal structure of the LINGO1 ectodomain shows that the LRRN3 protein takes on the characteristic horseshoe shape of most leucine-rich repeat proteins. The LINGO1 ectodomain also has a very long stretch of leucine-rich repeats which is the region that has the best alignment with the LRRN3 protein. This similar region shows that the LRRN3 protein has a structure that includes mostly beta-strands that are connected by loops, with a few alpha helices throughout.[14]
References
- GRCh38: Ensembl release 89: ENSG00000173114 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000036295 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: leucine rich repeat neuronal 3".
- Fukamachi K, Matsuoka Y, Kitanaka C, Kuchino Y, Tsuda H (September 2001). "Rat neuronal leucine-rich repeat protein-3: cloning and regulation of the gene expression". Biochem. Biophys. Res. Commun. 287 (1): 257–63. doi:10.1006/bbrc.2001.5579. PMID 11549284.
- "Gene Cards: Leucine-Rich Repeat Neuronal 3".
- "ACEView: "Homo sapiens" complex locus LRRN3".
- "ExPASY Proteomics Server". Archived from the original on 2003-07-23.
- "EMBL-EBI: Leucine-rich Repeat".
- "SDSC Biology Workbench: Multiple Sequence Alignment".
- "UniGene: Leucine-Rich Repeat Neuronal 3".
- "Allen Brain Atlas: Leucine-Rich Repeat Protein 3, Neuronal".
- "The Journal of Biological Chemistry: The Structure of the Lingo-1 Ectodomain".
Further reading
- Rose JE, Behm FM, Drgon T, et al. "Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score". Mol. Med. 16 (7–8): 247–53. doi:10.2119/molmed.2009.00159. PMC 2896464. PMID 20379614.
- Sousa I, Clark TG, Holt R, et al. (2010). "Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry". Mol Autism. 1 (1): 7. doi:10.1186/2040-2392-1-7. PMC 2913944. PMID 20678249.