I-CreI
I-CreI is a homing endonuclease whose gene was first discovered in the chloroplast genome of Chlamydomonas reinhardtii, a species of unicellular green algae.[1] It is named for the facts that: it resides in an Intron; it was isolated from Clamydomonas reinhardtii; it was the first (I) such gene isolated from C. reinhardtii. Its gene resides in a group I intron in the 23S ribosomal RNA gene of the C. reinhardtii chloroplast, and I-CreI is only expressed when its mRNA is spliced from the primary transcript of the 23S gene. I-CreI enzyme, which functions as a homodimer, recognizes a 22-nucleotide sequence of duplex DNA and cleaves one phosphodiester bond on each strand at specific positions. I-CreI is a member of the LAGLIDADG family of homing endonucleases, all of which have a conserved LAGLIDADG amino acid motif that contributes to their associative domains and active sites. When the I-CreI-containing intron encounters a 23S gene lacking the intron, I-CreI enzyme "homes" in on the "intron-minus" allele of 23S and effects its parent intron's insertion into the intron-minus allele. Introns with this behavior are called mobile introns. Because I-CreI provides for its own propagation while conferring no benefit on its host, it is an example of selfish DNA.
| DNA endonuclease I-CreI | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | ? | ||||||
| UniProt | P05725 | ||||||
| |||||||
Discovery
I-CreI was first observed as an intervening sequence in the 23S rRNA gene of the C. reinhardtii chloroplast genome.[1] The 23S gene is an RNA gene, meaning that its transcript is not translated into protein. As RNA, it forms part of the large subunit of the ribosome. An open reading frame coding for a 163-amino acid protein was found in this 23S intron, suggesting that a protein might facilitate the homing behavior of the mobile intron. Furthermore, the predicted protein had a LAGLIDADG motif, a conserved amino acid sequence that is present in other proteins coded for in group I mobile introns. A 1991 study established that the ORF codes for a DNA endonuclease, I-CreI, which selectively cuts a site corresponding to where the intron is spliced out of the 23S primary transcript.[2] The study also showed that the intron was able to invade 23S alleles that did not already have it.[2]
Mechanism of propagation
I-CreI has evolved to cut a 22-nucleotide sequence of DNA that occurs in alleles of the 23S ribosomal RNA gene that lack the I-CreI-containing intron. When such an "intron-minus" allele is cut, pathways of double-strand break repair are activated in the cell. The cell uses as a template for repair the 23S allele that yielded the responsible I-CreI enzyme, thus replicating the I-CreI-containing intron.[3] The resulting "intron-plus" allele no longer contains an intact homing site for the I-CreI enzyme, and is therefore not cleaved. Since this intron provides for its own replication without conferring any benefit on its host, I-CreI is a form of selfish DNA.
Structural studies and possible applications
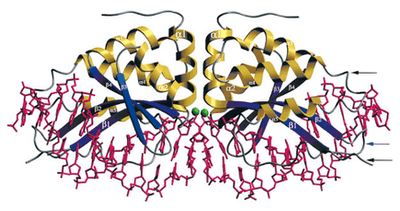
Because I-CreI has evolved to cut such a long sequence of DNA, unlike restriction endonucleases that typically cut four- or six-nucleotide sequences, it is capable of cutting a single site within a very large genome. A four- or six-nucleotide sequence is expected to occur many, many times in a genome of millions or billions of nucleotides simply by chance, whereas a 22-nucleotide sequence might occur only once (109/46 vs. 109/422). This specificity of I-CreI cleavage makes I-CreI a promising tool for gene targeting. If a person were to have a disease due to a defective allele of some gene, it would be helpful to be able to replace that allele with a functional one. If one could cause I-CreI to cut the DNA only in the defective allele while simultaneously providing a normal allele for the cell to use as a repair template, the patient's own homologous recombination machinery could insert the desired allele in place of the dysfunctional one. The specificity of I-CreI also allows for the reduction of deleterious effects due to double-strand breaks outside of the gene of interest.
In order to use I-CreI as a tool in this fashion, it is necessary to make it recognize and cleave sequences of DNA different from its native homing site. An Escherichia coli genetic system for studying the relationship between I-CreI structure and its homing site specificity was created in 1997.[5] In 1997, the structure of the I-CreI protein was determined,[6] and in 1998, its crystal structure bound to its native DNA homing site was solved, greatly aiding research in altering the homing site recognition of the protein.[4] Mutant forms of the protein have since been created that exhibit altered homing site specificity.[7][8][9] A genetic system in Saccharomyces cerevisiae has also been created, yielding additional I-CreI mutants with modified homing site specificities.[10][11]
I-CreI has already been used successfully to induce homologous recombination in Drosophila melanogaster, an extremely popular eukaryotic model organism.[12] It seems very likely that advances in molecular biological techniques and generation of a library of I-CreI-derived novel endonucleases will eventually allow for the targeting of many genes of etiological significance.
References
- Rochaix, JD; Malnoe, P (1978). "Anatomy of the chloroplast ribosomal DNA of Chlamydomonas reinhardtii". Cell. 15 (2): 661–670. doi:10.1016/0092-8674(78)90034-x. PMID 719757.
- Dürrenberger F, Rochaix JD (November 1991). "Chloroplast ribosomal intron of Chlamydomonas reinhardtii: in vitro self-splicing, DNA endonuclease activity and in vivo mobility". The EMBO Journal. 10 (11): 3495–501. doi:10.1002/j.1460-2075.1991.tb04913.x. PMC 453078. PMID 1915304.
- Dürrenberger F, Thompson AJ, Herrin DL, Rochaix JD (September 1996). "Double strand break-induced recombination in Chlamydomonas reinhardtii chloroplasts". Nucleic Acids Research. 24 (17): 3323–31. doi:10.1093/nar/24.17.3323. PMC 146090. PMID 8811085.
- Jurica MS, Monnat RJ, Stoddard BL (October 1998). "DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI". Molecular Cell. 2 (4): 469–76. doi:10.1016/s1097-2765(00)80146-x. PMID 9809068.
- Seligman, LM; Stephens, KM; Savage, JH; Monnat, RJ (1997). "Genetic Analysis of the Chlamydomonas reinhardtii I-CreI Mobile intron Homing System in Escherichia coli". Genetics. 147 (4): 1653–1664. PMC 1208338. PMID 9409828.
- Heath PJ, Stephens KM, Monnat RJ, Stoddard BL (June 1997). "The structure of I-Crel, a group I intron-encoded homing endonuclease". Nature Structural Biology. 4 (6): 468–76. doi:10.1038/nsb0697-468. PMID 9187655.
- Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL (September 2002). "Mutations altering the cleavage specificity of a homing endonuclease". Nucleic Acids Research. 30 (17): 3870–9. doi:10.1093/nar/gkf495. PMC 137417. PMID 12202772.
- Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Stoddard BL, Seligman LM (September 2004). "Isolation and characterization of new homing endonuclease specificities at individual target site positions". Journal of Molecular Biology. 342 (1): 31–41. doi:10.1016/j.jmb.2004.07.031. PMID 15313605.
- Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM (2006). "Homing endonuclease I-CreI derivatives with novel DNA target specificities". Nucleic Acids Research. 34 (17): 4791–800. doi:10.1093/nar/gkl645. PMC 1635285. PMID 16971456.
- Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, Rolland S, Prieto J, Blanco FJ, Bravo J, Montoya G, Serrano L, Duchateau P, Pâques F (January 2006). "Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets". Journal of Molecular Biology. 355 (3): 443–58. doi:10.1016/j.jmb.2005.10.065. PMID 16310802.
- Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, Montoya G, Pâques F, Duchateau P (2006). "A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences". Nucleic Acids Research. 34 (22): e149. doi:10.1093/nar/gkl720. PMC 1702487. PMID 17130168.
- Maggert KA, Golic KG (November 2005). "Highly efficient sex chromosome interchanges produced by I-CreI expression in Drosophila". Genetics. 171 (3): 1103–14. doi:10.1534/genetics.104.040071. PMC 1456814. PMID 16020774.