Hologenome theory of evolution
The hologenome theory of evolution[1][2][3][4] recasts the individual animal or plant (and other multicellular organisms) as a community or a "holobiont" – the host plus all of its symbiotic microbes. Consequently, the collective genomes of the holobiont form a "hologenome". Holobionts and hologenomes are structural entities[5] that replace misnomers in the context of host-microbiota symbioses such as superorganism (i.e., an integrated social unit composed of conspecifics), organ, and metagenome. Variation in the hologenome may encode phenotypic plasticity of the holobiont and can be subject to evolutionary changes caused by selection and drift, if portions of the hologenome are transmitted between generations with reasonable fidelity. One of the important outcomes of recasting the individual as a holobiont subject to evolutionary forces is that genetic variation in the hologenome can be brought about by changes in the host genome and also by changes in the microbiome, including new acquisitions of microbes, horizontal gene transfers, and changes in microbial abundance within hosts. Although there is a rich literature on binary host–microbe symbioses, the hologenome concept distinguishes itself by including the vast symbiotic complexity inherent in many multicellular hosts. For recent literature on holobionts and hologenomes published in an open access platform, see the following reference.[4]
Origin
Lynn Margulis coined the term holobiont in her 1991 book Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis (MIT Press)[6], though this was not in the context of diverse populations of microbes. The term holobiont is derived from the Ancient Greek ὅλος (hólos, "whole"), and the word biont for a unit of life.[7]
In September 1994, Richard Jefferson coined the term hologenome when he introduced the hologenome theory of evolution at a presentation at Cold Spring Harbor Laboratory.[1][8][9] At the CSH Symposium and earlier, the unsettling number and diversity of microbes that were being discovered through the powerful tool of PCR-amplification of 16S ribosomal RNA genes was exciting, but confusing interpretations in diverse studies. A number of speakers referred to microbial contributions to mammalian or plant DNA samples as 'contamination'. In his lecture, Jefferson argued that these were likely not contamination, but rather essential components of the samples that reflected the actual genetic composition of the organism being studied, integral to the complex system in which it lives. This implied that the logic of the organism's performance and capabilities would be embedded only in the hologenome. Observations on the ubiquity of microbes in plant and soil samples as well as laboratory work on molecular genetics of vertebrate-associated microbial enzymes impacting hormone action informed this hypothesis[10]. References was made to work indicating that mating pheromones were only released after skin microbiota activated the precursors.[11]
At the 14th South African Congress of Biochemistry and Molecular Biology in 1997,[12] Jefferson described how the modulation of steroid and other hormone levels by microbial glucuronidases and arylsulfatase profoundly impacted the performance of the composite entity. Following on work done isolating numerous and diverse glucuronidases from microbial samples of African animal feces,[13] and their differential cleavage of hormones, he hypothesized that this phenomenon, microbially-mediated hormone modulation, could underlie evolution of disease and social behavior as well as the holobiont fitness and system resilience. In his lectures, Jefferson coined and defined the term ‘Ecotherapeutics’, referring to adjustment of the population structure of the microbial composition in plants and animals - the microbiome - and their support ecosystem to improve performance.[9][12] In 2007, Jefferson followed with a series of posts on the logic of hologenome theory on Cambia's Science as Social Enterprise page.[14]
In 2008, Eugene Rosenberg and Ilana Zilber-Rosenberg apparently independently used the term hologenome and developed the hologenome theory of evolution.[15] This theory was originally based on their observations of Vibrio shiloi-mediated bleaching of the coral Oculina patagonica. Since its first introduction, the theory has been promoted as a fusion of Lamarckism and Darwinism and expanded to all of evolution, not just that of corals. The history of the development of the hologenome theory and the logic undergirding its development was the focus of a cover article by Carrie Arnold in New Scientist in January, 2013.[16] A comprehensive treatment of the theory, including updates by the Rosenbergs on neutrality, pathogenesis and multi-level selection, can be found in their 2013 book.[2]
In 2013, Robert Brucker and Seth Bordenstein[17] re-invigorated the hologenome concept by showing that the gut microbiomes of closely related Nasonia wasp species are distinguishable, and contribute to hybrid death. This set interactions between hosts and microbes in a conceptual continuum with interactions between genes in the same genome. In 2015, Bordenstein and Kevin R. Theis outlined a conceptual framework that aligns with pre-existing theories in biology.[4]
Support from vertebrate biology
Multicellular life is made possible by the coordination of physically and temporally distinct processes, most prominently through hormones. Hormones mediate critical activities in vertebrates, including ontogeny, somatic and reproductive physiology, sexual development, performance and behaviour.
Many of these hormones – including most steroids and thyroxines – are secreted in inactive form through the endocrine and apocrine systems into epithelial corridors in which microbiota are widespread and diverse, including gut, urinary tract, lung and skin. There, the inactive hormones can be re-activated by cleavage of the glucuronide or sulfate residue, allowing them to be reabsorbed. Thus the concentration and bioavailability of many of the hormones is impacted by microbial cleavage of conjugated intermediaries, itself determined by a diverse population with redundant enzymatic capabilities. Aspects of enterohepatic circulation have been known for decades, but had been viewed as an ancillary effect of detoxification and excretion of metabolites and xenobiotics, including effects on lifetimes of pharmaceuticals, including birth control formulations.
The basic premise of Jefferson's first exposition of the hologenome theory is that a spectrum of hormones can be re-activated and resorbed from epithelia, potentially modulating effective time and dose relationships of many vertebrate hormones. The ability to alter and modulate, amplify and suppress, disseminate and recruit new capabilities as microbially-encoded 'traits' means that sampling, sensing and responding to the environment become intrinsic features and emergent capabilities of the holobiont, with mechanisms that can provide rapid, sensitive, nuanced and persistent performance changes.
Studies by Froebe et al.[18] in 1990 indicating that essential mating pheromones, including androstenols, required activation by skin-associated microbial glucuronidases and sulfatases. In the absence of microbial populations in the skin, no detectable aromatic pheromone was released, as the pro-pheromone remained water-soluble and non-volatile. This effectively meant that the microbes in the skin were essential to produce a mating signal.[19]
Support from coral biology
Subsequent re-articulation describing the hologenome theory by Rosenberg and Zilber-Rosenberg, published 13 years after Jefferson's definition of the theory, was based on their observations of corals, and the coral probiotic hypothesis.
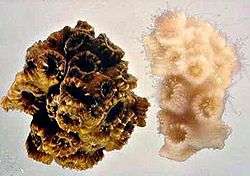
Coral reefs are the largest structures created by living organisms, and contain abundant and highly complex microbial communities. A coral "head" is a colony of genetically identical polyps, which secrete an exoskeleton near the base. Depending on the species, the exoskeleton may be hard, based on calcium carbonate, or soft and proteinaceous. Over many generations, the colony creates a large skeleton that is characteristic of the species. Diverse forms of life take up residence in a coral colony, including photosynthetic algae such as Symbiodinium, as well as a wide range of bacteria including nitrogen fixers,[20] and chitin decomposers,[21] all of which form an important part of coral nutrition.[22] The association between coral and its microbiota is species dependent, and different bacterial populations are found in mucus, skeleton and tissue from the same coral fragment.[23]
Over the past several decades, major declines in coral populations have occurred. Climate change, water pollution and overfishing are three stress factors that have been described as leading to disease susceptibility. Over twenty different coral diseases have been described, but of these, only a handful have had their causative agents isolated and characterized.
Coral bleaching is the most serious of these diseases. In the Mediterranean Sea, the bleaching of Oculina patagonica was first described in 1994 and, through a rigorous application of Koch's Postulates, determined to be due to infection by Vibrio shiloi.[24] From 1994 to 2002, bacterial bleaching of O. patagonica occurred every summer in the eastern Mediterranean. Surprisingly, however, after 2003, O. patagonica in the eastern Mediterranean has been resistant to V. shiloi infection, although other diseases still cause bleaching.
The surprise stems from the knowledge that corals are long lived, with lifespans on the order of decades,[25] and do not have adaptive immune systems. Their innate immune systems do not produce antibodies, and they should seemingly not be able to respond to new challenges except over evolutionary time scales. Yet multiple researchers have documented variations in bleaching susceptibility that may be termed 'experience-mediated tolerance'.[26][27] The puzzle of how corals managed to acquire resistance to a specific pathogen led Eugene Rosenberg and Ilana Zilber-Rosenberg to propose the Coral Probiotic Hypothesis.[23] This hypothesis proposes that a dynamic relationship exists between corals and their symbiotic microbial communities. Beneficial mutations can arise and spread among the symbiotic microbes much faster than in the host corals. By altering its microbial composition, the "holobiont" can adapt to changing environmental conditions far more rapidly than by genetic mutation and selection in the host species alone.
Extrapolating the coral probiotic hypothesis to other organisms, including higher plants and animals, led to the Rosenberg's support for and publications around the hologenome theory of evolution.
Theory
Definition
The framework of the hologenome theory of evolution is as follows (condensed from Rosenberg et al., 2007):[28]
- "All animals and plants establish symbiotic relationships with microorganisms."
- "Different host species contain different symbiont populations and individuals of the same species can also contain different symbiont populations."
- "The association between a host organism and its microbial community affect both the host and its microbiota."
- "The genetic information encoded by microorganisms can change under environmental demands more rapidly, and by more processes, than the genetic information encoded by the host organism."
- "... the genome of the host can act in consortium with the genomes of the associated symbiotic microorganisms to create a hologenome. This hologenome...can change more rapidly than the host genome alone, thereby conferring greater adaptive potential to the combined holobiont evolution."
- "Each of these points taken together [led Rosenberg et al. to propose that] the holobiont with its hologenome should be considered as the unit of natural selection in evolution."
Some authors supplement the above principles with an additional one. If a given holobiont is to be considered a unit of natural selection:
Ten principles of holobionts and hologenomes were presented in PLOS Biology:[4]
- I. Holobionts and hologenomes are units of biological organization
- II. Holobionts and hologenomes are not organ systems, superorganisms, or metagenomes
- III. The hologenome is a comprehensive gene system
- IV. The hologenome concept reboots elements of Lamarckian evolution
- V. Hologenomic variation integrates all mechanisms of mutation
- VI. Hologenomic evolution is most easily understood by equating a gene in the nuclear genome to a microbe in the microbiome
- VII. The hologenome concept fits squarely into genetics and accommodates multilevel selection theory
- VIII. The hologenome is shaped by selection and neutrality
- IX. Hologenomic speciation blends genetics and symbiosis
- X. Holobionts and their hologenomes do not change the rules of evolutionary biology
Horizontally versus vertically transmitted symbionts
Many case studies clearly demonstrate the importance of an organism's associated microbiota to its existence. (For example, see the numerous case studies in the Microbiome article.) However, horizontal versus vertical transmission of endosymbionts must be distinguished. Endosymbionts whose transmission is predominantly vertical may be considered as contributing to the heritable genetic variation present in a host species.[29]
In the case of colonial organisms such as corals, the microbial associations of the colony persist even though individual members of the colony, reproducing asexually, live and die. Corals also have a sexual mode of reproduction, resulting in planktonic larva; it is less clear whether microbial associations persist through this stage of growth. Also, the bacterial community of a colony may change with the seasons.[23]
Many insects maintain heritable obligate symbiosis relationships with bacterial partners. For example, normal development of female wasps of the species Asobara tabida is dependent on Wolbachia infection. If "cured" of the infection, their ovaries degenerate.[31] Transmission of the infection is vertical through the egg cytoplasm.
In contrast, many obligate symbiosis relationships have been described in the literature where transmission of the symbionts is via horizontal transfer. A well-studied example is the nocturnally feeding squid Euprymna scolopes, which camouflages its outline against the moonlit ocean surface by emitting light from its underside with the aid of the symbiotic bacterium Vibrio fischeri.[32] The Rosenbergs cite this example within the context of the hologenome theory of evolution.[33] Squid and bacterium maintain a highly co-evolved relationship. The newly hatched squid collects its bacteria from the sea water, and lateral transfer of symbionts between hosts permits faster transfer of beneficial mutations within a host species than are possible with mutations within the host genome.
Primary versus secondary symbionts

Another traditional distinction between endosymbionts has been between primary and secondary symbionts.[29] Primary endosymbionts reside in specialized host cells that may be organized into larger, organ-like structures (in insects, the bacteriome). Associations between hosts and primary endosymbionts are usually ancient, with an estimated age of tens to hundreds of millions of years. According to endosymbiotic theory, extreme cases of primary endosymbionts include mitochondria, plastids (including chloroplasts), and possibly other organelles of eukaryotic cells. Primary endosymbionts are usually transmitted exclusively vertically, and the relationship is always mutualistic and generally obligate for both partners. Primary endosymbiosis is surprisingly common. An estimated 15% of insect species, for example, harbor this type of endosymbiont.[34] In contrast, secondary endosymbiosis is often facultative, at least from the host point of view, and the associations are less ancient. Secondary endosymbionts do not reside in specialized host tissues, but may dwell in the body cavity dispersed in fat, muscle, or nervous tissue, or may grow within the gut. Transmission may be via vertical, horizontal, or both vertical and horizontal transfer. The relationship between host and secondary endosymbiont is not necessarily beneficial to the host; indeed, the relationship may be parasitic.[29]
The distinction between vertical and horizontal transfer, and between primary and secondary endosymbiosis is not absolute, but follows a continuum, and may be subject to environmental influences. For example, in the stink bug Nezara viridula, the vertical transmission rate of symbionts, which females provide to offspring by smearing the eggs with gastric caeca, was 100% at 20 °C, but decreased to 8% at 30 °C.[35] Likewise, in aphids, the vertical transmission of bacteriocytes containing the primary endosymbiont Buchnera is drastically reduced at high temperature.[36] In like manner, the distinction between commensal, mutualistic, and parasitic relationships is also not absolute. An example is the relationship between legumes and rhizobial species: N2 uptake is energetically more costly than the uptake of fixed nitrogen from the soil, so soil N is preferred if not limiting. During the early stages of nodule formation, the plant-rhizobial relationship actually resembles a pathogenesis more than it does a mutualistic association.
Neo-Lamarckism within a Darwinian context
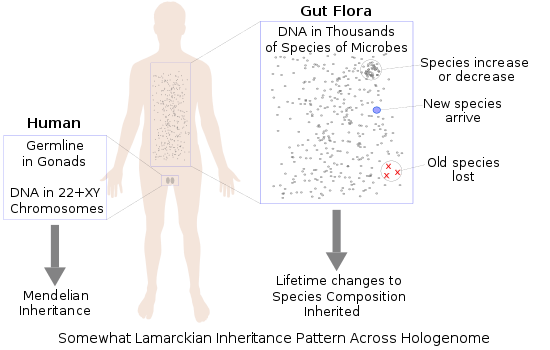
Lamarckism, the concept that an organism can pass on characteristics that it acquired during its lifetime to its offspring (also known as inheritance of acquired characteristics or soft inheritance) incorporated two common ideas of its time:
- Use and disuse – individuals lose characteristics they do not require (or use) and develop characteristics that are useful.
- Inheritance of acquired traits – individuals inherit the traits of their ancestors.
Although Lamarckian theory was rejected by the neo-Darwinism of the modern evolutionary synthesis in which evolution occurs through random variations being subject to natural selection, the hologenome theory has aspects that harken back to Lamarckian concepts. In addition to the traditionally recognized modes of variation (i.e. sexual recombination, chromosomal rearrangement, mutation), the holobiont allows for two additional mechanisms of variation that are specific to the hologenome theory: (1) changes in the relative population of existing microorganisms (i.e. amplification and reduction) and (2) acquisition of novel strains from the environment, which may be passed on to offspring.[33]
Changes in the relative population of existing microorganisms corresponds to Lamarckian "use and disuse", while the ability to acquire novel strains from the environment, which may be passed on to offspring, corresponds to Lamarckian "inheritance of acquired traits". The hologenome theory, therefore, is said by its proponents to incorporate Lamarckian aspects within a Darwinian framework.[33]
Additional case studies
-PLoS.jpg)
The pea aphid Acyrthosiphon pisum maintains an obligate symbiotic relationship with the bacterium Buchnera aphidicola, which is transmitted maternally to the embryos that develop within the mother's ovarioles. Pea aphids live on sap, which is rich in sugars but deficient in amino acids. They rely on their Buchnera endosymbiotic population for essential amino acids, supplying in exchange nutrients as well as a protected intracellular environment that allows Buchnera to grow and reproduce.[37] The relationship is actually more complicated than mutual nutrition; some strains of Buchnera increases host thermotolerance, while other strains do not. Both strains are present in field populations, suggesting that under some conditions, increased heat tolerance is advantageous to the host, while under other conditions, decreased heat tolerance but increased cold tolerance may be advantageous.[38] One can consider the variant Buchnera genomes as alleles for the larger hologenome.[30] The association between Buchnera and aphids began about 200 million years ago, with host and symbiont co-evolving since that time; in particular, it has been discovered that genome size in various Buchnera species has become extremely reduced, in some cases down to 450 kb, which is far smaller even than the 580 kb genome of Mycoplasma genitalium.[39]
Development of mating preferences, i.e. sexual selection, is considered to be an early event in speciation. In 1989, Dodd reported mating preferences in Drosophila that were induced by diet.[40] It has recently been demonstrated that when otherwise identical populations of Drosophila were switched in diet between molasses medium and starch medium, that the "molasses flies" preferred to mate with other molasses flies, while the "starch flies" preferred to mate with other starch flies. This mating preference appeared after only one generation and was maintained for at least 37 generations. The origin of these differences were changes in the flies' populations of a particular bacterial symbiont, Lactobacillus plantarum. Antibiotic treatment abolished the induced mating preferences. It has been suggested that the symbiotic bacteria changed the levels of cuticular hydrocarbon sex pheromones [41], however several other research papers have been unable to replicate this effect[42][43][44].
Zilber-Rosenberg and Rosenberg (2008) have tabulated many of the ways in which symbionts are transmitted and their contributions to the fitness of the holobiont, beginning with mitochondria found in all eukaryotes, chloroplast in plants, and then various associations described in specific systems. The microbial contributions to host fitness included provision of specific amino acids, growth at high temperatures, provision of nutritional needs from cellulose, nitrogen metabolism, recognition signals, more efficient food utilization, protection of eggs and embryos against metabolism, camouflage against predators, photosynthesis, breakdown of complex polymers, stimulation of the immune system, angiogenesis, vitamin synthesis, fiber breakdown, fat storage, supply of minerals from the soil, supply of organics, acceleration of mineralization, carbon cycling, and salt tolerance.[45]
Criticism
The hologenome theory is debated.[46] A major criticism by Ainsworth et al. has been their claim that V. shiloi was misidentified as the causative agent of coral bleaching, and that its presence in bleached O. patagonica was simply that of opportunistic colonization.[47]
If this is true, the original observation that led to Rosenberg's later articulation of the theory would be invalid. On the other hand, Ainsworth et al.[47] performed their samplings in 2005, two years after the Rosenberg group discovered O. patagonica no longer to be susceptible to V. shiloi infection; therefore their finding that bacteria are not the primary cause of present-day bleaching in Mediterranean coral O. patagonica should not be considered surprising. The rigorous satisfaction of Koch's postulates, as employed in Kushmaro et al. (1997),[24] is generally accepted as providing a definitive identification of infectious disease agents.
Baird et al. (2009)[25] have questioned basic assumptions made by Reshef et al. (2006)[23] in presuming that (1) coral generation times are too slow to adjust to novel stresses over the observed time scales, and that (2) the scale of dispersal of coral larvae is too large to allow for adaptation to local environments. They may simply have underestimated the potential rapidity of conventional means of natural selection. In cases of severe stress, multiple cases have been documented of ecologically significant evolutionary change occurring over a handful of generations.[48] Novel adaptive mechanisms such as switching symbionts might not be necessary for corals to adjust to rapid climate change or novel stressors.[25]
Organisms in symbiotic relationships evolve to accommodate each other, and the symbiotic relationship increases the overall fitness of the participant species. Although the hologenome theory is still being debated, it has gained a significant degree of popularity within the scientific community as a way of explaining rapid adaptive changes that are difficult to accommodate within a traditional Darwinian framework.[33]
Definitions and uses of the words holobiont and hologenome also differ between proponents and skeptics,[5] and the misuse of the terms has led to confusions over what comprises evidence related to the hologenome. Ongoing discourse is attempting to clear this confusion. Theis et al. clarify that "critiquing the hologenome concept is not synonymous with critiquing coevolution, and arguing that an entity is not a primary unit of selection dismisses the fact that the hologenome concept has always embraced multilevel selection."[5]
For instance,[49] Chandler and Turelli (2014) criticize the conclusions of Brucker and Bordenstein (2013), noting that their observations are also consistent with an alternative explanation. Brucker and Bordenstein (2014) responded to these criticisms, claiming they were unfounded[50] because of factual inaccuracies and altered arguments and definitions that were not advanced by Brucker and Bordenstein (2013).
Recently, Forest L Rohwer and colleagues developed a novel statistical test to examine the potential for the hologenome theory of evolution in coral species.[51] They found that coral species do not inherit microbial communities, and are instead colonized by a core group of microbes that associate with a diversity of species. The authors conclude: "Identification of these two symbiont communities supports the holobiont model and calls into question the hologenome theory of evolution." However, other studies in coral adhere to the original and pluralistic definitions of holobionts and hologenomes.[52] David Bourne, Kathleen Morrow and Nicole Webster clarify that "The combined genomes of this coral holobiont form a coral hologenome, and genomic interactions within the hologenome ultimately define the coral phenotype."
References
- Jefferson, Richard (2019-04-04). "Agriculture and the Third World". figshare. doi:10.6084/m9.figshare.7945781. Retrieved 2019-04-04.
- "The Hologenome Concept: Human, Animal and Plant Microbiota"
- Rosenberg, Eugene; Zilber-Rosenberg, Ilana (2016-05-04). "Microbes Drive Evolution of Animals and Plants: the Hologenome Concept". mBio. 7 (2): e01395–15. doi:10.1128/mBio.01395-15. ISSN 2150-7511. PMC 4817260. PMID 27034283.
- Bordenstein, Seth R.; Theis, Kevin R. (2015-08-18). "Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes". PLOS Biol. 13 (8): e1002226. doi:10.1371/journal.pbio.1002226. PMC 4540581. PMID 26284777.
- Theis, Kevin R.; Dheilly, Nolwenn M.; Klassen, Jonathan L.; Brucker, Robert M.; Baines, John F.; Bosch, Thomas C. G.; Cryan, John F.; Gilbert, Scott F.; Goodnight, Charles J. (2016-04-26). "Getting the Hologenome Concept Right: an Eco-Evolutionary Framework for Hosts and Their Microbiomes". mSystems. 1 (2): e00028–16. doi:10.1128/mSystems.00028-16. ISSN 2379-5077. PMC 5069740. PMID 27822520.
- Margulis, Lynn; Fester, René (1991-01-01). Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. MIT Press. ISBN 9780262132695.
- Suárez, Javier (2018-10-01). "The importance of symbiosis in philosophy of biology: an analysis of the current debate on biological individuality and its historical roots". Symbiosis. 76 (2): 77–96. doi:10.1007/s13199-018-0556-1. ISSN 1878-7665.
- Number 6 in a series of 7 VHS recordings, A Decade of PCR: Celebrating 10 Years of Amplification, Cold Spring Harbor Laboratory Press, 1994. ISBN 0-87969-473-4. http://www.cshlpress.com/default.tpl?..
- Part 4: The Hologenome - YouTube Agriculture, Environment and the Developing World: A Future of PCR. Part 4: The Hologenome Plenary lecture by Richard Jefferson, CEO Cambia, Cold Spring Harbor, September, 1994.
- Wilson, Kate J.; Giller, Ken E.; Jefferson, Richard A. (1991), Hennecke, Hauke; Verma, Desh Pal S. (eds.), "β-Glucuronidase (GUS) Operon Fusions as a Tool for Studying Plant-Microbe Interactions", Advances in Molecular Genetics of Plant-Microbe Interactions Vol. 1: Proceedings of the 5th International Symposium on the Molecular Genetics of Plant-Microbe Interactions, Interlaken, Switzerland, September 9–14, 1990, Current Plant Science and Biotechnology in Agriculture, Springer Netherlands, pp. 226–229, doi:10.1007/978-94-015-7934-6_35, ISBN 9789401579346
- "Inhibitor of odor-producing axillary bacterial exoenzymes - Abstract". journal.scconline.org. Retrieved 2019-03-22.
- Jefferson, Richard A; Wilson, Kate J; Liang, Weijun. (1997-01-15). "Molecular genetics of the E. coli gus operon: Medical and evolutionary implications for glucuronide and xenobiotic metabolism". The 14th Congress of the South African Society of Biochemistry and Molecular Biology, Grahamstown, South Africa. doi:10.6084/m9.figshare.7951037.
- "Early Guses and Repressor - 2 Patent Records". The Lens - Free & Open Patent and Scholarly Search. Retrieved 2019-04-03.
- Jefferson, Richard A. "Science as Social Enterprise - Thoughts from Richard Jefferson and Cambia on democratizing science-enabled innovation". blogs.cambia.org. Retrieved 2019-04-03.
- Zilber-Rosenberg, Ilana (2008). "Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution". FEMS Microbiology Reviews. 32 (5): 723–35. doi:10.1111/j.1574-6976.2008.00123.x. PMID 18549407.
- "Creator of Species, The Hologenome, a New View of Evolution" https://www.newscientist.com/article/mg21728992-000-the-hologenome-a-new-view-of-evolution/
- Brucker, Robert M; Bordenstein, Seth R (2013). "The Hologenomic Basis of Speciation: Gut Bacteria Cause Hybrid Lethality in the Genus Nasonia". Science. 341 (6146): 667–669. Bibcode:2013Sci...341..667B. doi:10.1126/science.1240659. PMID 23868918.
- "Axillary malodor production: A new mechanism" (PDF). Archived from the original (PDF) on 2015-07-24. Retrieved 2015-08-07.
- Enabling Innovation: A 2011 Lecture in the Illahee Series https://vimeo.com/26387884
- Shashar N, Feldstein T, Cohen Y, Loya Y (1994). "Nitrogen fixation (acetylene reduction) on a coral reef". Coral Reefs. 13 (3): 171–4. Bibcode:1994CorRe..13..171S. doi:10.1007/BF00301195.
- Ducklow HW, Mitchell R (1979). "Bacterial Populations and Adaptations in the Mucus Layers on Living Corals". Limnology and Oceanography. 24 (4): 715–725. Bibcode:1979LimOc..24..715D. doi:10.4319/lo.1979.24.4.0715. JSTOR 2835723.
- Kushmaro A, Kramarsky-Winter E (2004). "Bacteria as a source of coral nutrition". In Rosenberg E, Loya Y (eds.). Coral Health and Disease. Berlin Heidelberg: Springer-Verlag. pp. 231–241. ISBN 978-3-540-20772-6.
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E (2006). "The Coral Probiotic Hypothesis" (PDF). Environmental Microbiology. 8 (12): 2068–73. CiteSeerX 10.1.1.627.6120. doi:10.1111/j.1462-2920.2006.01148.x. PMID 17107548.
- Kushmaro A, Rosenberg E, Fine M, Loya Y (1997). "Bleaching of the coral Oculina patagonica by Vibrio AK-1". Marine Ecology Progress Series. 147 (1): 159–165. Bibcode:1997MEPS..147..159K. doi:10.3354/meps147159. Archived from the original on 2013-01-29.
The causative agent, Vibrio AK-1, was present in 28 bleached O. patagonica examined, but absent from 24 healthy (unbleached) corals. The Vibrio sp. was isolated in pure culture, characterized microbiologically, and shown to cause bleaching when inoculated onto unbleached corals.
- Baird AH, Bhagooli R, Ralph PJ, Takahashi S (2009). "Coral bleaching: the role of the host" (PDF). Trends in Ecology and Evolution. 24 (1): 16–20. doi:10.1016/j.tree.2008.09.005. PMID 19022522.
- Brown B, Dunne R, Goodson M, Douglas A (2002). "Experience shapes the susceptibility of a reef coral to bleaching". Coral Reefs. 21: 119–126. doi:10.1007/s00338-002-0215-z.
- Richardson LL, Aronson RB (2000). "Infectious diseases of reef corals" (PDF). Proceedings 9th International Coral Reef Symposium. 2: 1225–30. Archived from the original (PDF) on 2015-09-23. Retrieved 2012-03-01.
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007). "The role of microorganisms in coral health, disease and evolution" (PDF). Nature Reviews Microbiology. 5 (5): 355–362. doi:10.1038/nrmicro1635. PMID 17384666.
- Feldhaar H (2011). "Bacterial symbionts as mediators of ecologically important traits of insect hosts". Ecological Entomology. 36 (5): 533–543. doi:10.1111/j.1365-2311.2011.01318.x.
- Gilbert SF, McDonald E, Boyle N, Buttino N, Gyi L, Mai M, Prakash N, Robinson J (2010). "Symbiosis as a source of selectable epigenetic variation: taking the heat for the big guy". Phil. Trans. R. Soc. B. 365 (1540): 671–678. doi:10.1098/rstb.2009.0245. PMC 2817139. PMID 20083641.
- Pannebakker BA, Loppin B, Elemans CP, Humblot L, Vavre F (2007). "Parasitic inhibition of cell death facilitates symbiosis". Proc. Natl. Acad. Sci. USA. 104 (1): 213–5. Bibcode:2007PNAS..104..213P. doi:10.1073/pnas.0607845104. PMC 1765438. PMID 17190825.
- Visick KL, Ruby EG (2006). "Vibrio fischeri and its host: it takes two to tango" (PDF). Current Opinion in Microbiology. 9 (6): 632–8. doi:10.1016/j.mib.2006.10.001. PMID 17049299. Archived from the original (PDF) on 2010-06-11.
- Rosenberg E, Zilber-Rosenberg I (2011). "Symbiosis and development: The hologenome concept". Birth Defects Research Part C: Embryo Today: Reviews. 93 (1): 56–66. doi:10.1002/bdrc.20196. PMID 21425442.
- Baumann P (2005). "Bacteriology of bacteriocyte-associated endosymbionts of plant sap-sucking insects". Annual Review of Microbiology. 59: 155–189. doi:10.1146/annurev.micro.59.030804.121041. PMID 16153167.
- Prado SS, Golden M, Follett PA, Daugherty MP, Almeida RP (2009). "Demography of Gut Symbiotic and Aposymbiotic Nezara viridula L. (Hemiptera: Pentatomidae)" (PDF). Environmental Entomology. 38 (1): 103–109. doi:10.1603/022.038.0112. PMID 19791602.
- Montllor CB, Maxmen A, Purcell AH (2002). "Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress". Ecological Entomology. 27 (2): 189–195. doi:10.1046/j.1365-2311.2002.00393.x.
- Gómez-Valero L, Soriano-Navarro M, Pérez-Brocal V, Heddi A, Moya A, García-Verdugo JM, Latorre A (2004). "Coexistence of Wolbachia with Buchnera aphidicola and a Secondary Symbiont in the Aphid Cinara cedri". J. Bacteriol. 186 (19): 6626–33. doi:10.1128/JB.186.19.6626-6633.2004. PMC 516615. PMID 15375144.
- Dunbar HE, Wilson AC, Ferguson NR, Moran NA (May 2007). "Aphid Thermal Tolerance Is Governed by a Point Mutation in Bacterial Symbionts". PLoS Biology. 5 (5): e96. doi:10.1371/journal.pbio.0050096. PMC 1847839. PMID 17425405.
- Gil R, Sabater-Muñoz B, Latorre A, Silva FJ, Moya A (April 2002). "Extreme genome reduction in Buchnera spp.: Toward the minimal genome needed for symbiotic life". Proc. Natl. Acad. Sci. USA. 99 (7): 4454–8. Bibcode:2002PNAS...99.4454G. doi:10.1073/pnas.062067299. PMC 123669. PMID 11904373.
- Dodd DMB (September 1989). "Reproductive Isolation as a Consequence of Adaptive Divergence in Drosophila pseudoobscura" (PDF). Evolution. 43 (6): 1308–11. doi:10.2307/2409365. JSTOR 2409365. PMID 28564510. Archived from the original (PDF) on 2011-03-02.
- Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E (November 2010). "Commensal bacteria play a role in mating preference of Drosophila melanogaster". Proc. Natl. Acad. Sci. USA. 107 (46): 20051–6. Bibcode:2010PNAS..10720051S. doi:10.1073/pnas.1009906107. PMC 2993361. PMID 21041648.
- Najarro, Michael A.; Sumethasorn, Matt; Lamoureux, Alexandra; Turner, Thomas L. (2015-08-06). "Choosing mates based on the diet of your ancestors: replication of non-genetic assortative mating in Drosophila melanogaster". PeerJ. 3: e1173. doi:10.7717/peerj.1173. ISSN 2167-8359.
- Leftwich, Philip T.; Hutchings, Matthew I.; Chapman, Tracey (2018). "Diet, Gut Microbes and Host Mate Choice". BioEssays. 40 (12): 1800053. doi:10.1002/bies.201800053. ISSN 1521-1878.
- Leftwich, Philip T.; Clarke, Naomi V. E.; Hutchings, Matthew I.; Chapman, Tracey (2017-11-28). "Gut microbiomes and reproductive isolation in Drosophila". Proceedings of the National Academy of Sciences. 114 (48): 12767–12772. doi:10.1073/pnas.1708345114. ISSN 0027-8424. PMC 5715749. PMID 29109277.
- Zilber-Rosenberg I, Rosenberg E (2008). "Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution". FEMS Microbiology Reviews. 32 (5): 723–735. doi:10.1111/j.1574-6976.2008.00123.x. PMID 18549407.
- Leggat W, Ainsworth T, Bythell J, Dove S, Gates R, Hoegh-Guldberg O, Iglesias-Prieto R, Yellowlees D (2007). "The hologenome theory disregards the coral holobiont". Nature Reviews Microbiology. 5 (10): Online Correspondence. doi:10.1038/nrmicro1635-c1.
- Ainsworth TD, Fine M, Roff G, Hoegh-Guldberg O (2008). "Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica". The ISME Journal. 2 (1): 67–73. doi:10.1038/ismej.2007.88. PMID 18059488.
- Carroll SP, Hendry AP, Reznick DN, Fox CW (2007). "Evolution on ecological time-scales". Functional Ecology. 21 (3): 387–393. doi:10.1111/j.1365-2435.2007.01289.x.
- Comment on “The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia” | Science
- Response to Comment on “The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia” | Science
- The ISME Journal - Stable and sporadic symbiotic communities of coral and algal holobionts
- Bourne, David G.; Morrow, Kathleen M.; Webster, Nicole S. (2016-01-01). "Insights into the Coral Microbiome: Underpinning the Health and Resilience of Reef Ecosystems". Annual Review of Microbiology. 70 (1): 317–40. doi:10.1146/annurev-micro-102215-095440. PMID 27482741.