Glutamate rich 5
Glutamate Rich Protein 5 is a protein in humans encoded by the ERICH5 gene, also known as Chromosome 8 open reading frame 47 (C8orf47).
| ERICH5 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | ERICH5, C8orf47, glutamate rich 5 | ||||||||||||||||||||||||
| External IDs | MGI: 2447772 HomoloGene: 52129 GeneCards: ERICH5 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 8: 98.06 – 98.09 Mb | Chr 15: 34.45 – 34.47 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Gene
The ERICH5 gene is located on human chromosome 8 at 8q22.2 and spans 29 kb on the plus strand of the DNA. [5] ERICH5 contains three exons and two introns. [6] ERICH5 is also known as C8orf47 and Glutamate Rich 5.[5]
Protein
Isoforms
The ERICH5 protein has two isofroms. The longest isofrom, isofrom 1, spans 1,550 base pairs and is composed of 374 amino acids. The second isoform lacks the third and final exon and is 596 base pairs long.[6]
Domains and motifs
ERICH5 contains one conserved domain, a domain of unknown function called DUF4573.[7] ERICH5 is predicted to contain two highly conserved motifs, an APC/C binding motif and the LIG_FHA_2 motif.[8] The APC/C motif spans amino acid 222-226 and serves as a binding site for the Anaphase- promoting complex.[9] The LIG_FHA_2 motif is involved in the cell checkpoint pathway and is found in many proteins localized in the nucleus that regulate cell cycle.[10]
Post-translational modifications
ERICH5 is predicted to undergo several post translational modifications including phosphorylation, O-glycosylation, and Sumolyation. [11][12] [13]Many of the phosphorylation sites and O-glycosylation sites were predicted at the same amino acid. The post translational modifications shown are those conserved among ERICH5 orthologs. Several kinases were predicted to phosphorylate ERICH5 including PKC, cdc2, CKI, PKA, DNAPK, ATM, EGFR, and CKII.[11]
| Post Translational Modification | Amino Acid Location |
| Phosphorylation | S4, S5, S27, S32, S49, T50, S58, S100, T101, T104, T138, S169, S227, S234, T239,T248, S274, T289, S307,Y320, T340, T346 |
| O-Glycosylation | S5,S27, S32, S49, T50, S58, S100, T101, T104, T138, S169, S227, S234, T239, T248, S274, T289,S307 |
| Sumolyation | K121, K131, K211, K251, ,K343, K360 |
Secondary structure
ERICH5 was predicted to contain three alpha helices and two beta sheets as well as regions of random coils. [14]
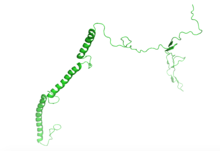
Sub-cellular localization
ERICH5 was predicted to be localized in the nucleus.[16]
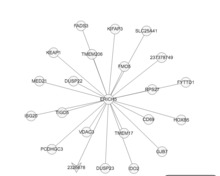
Protein interactions
ERICH5 was predicted to interact with several proteins through yeast two hybrid screening and affinity chromatography.[15] Several of the proteins ERICH5 was predicted to interact with were also localized in the nucleus.[15]
Expression
Normal expression
ERICH5 shows elevated levels of expression in the fetal liver, liver, pancreas, and retina compared to other tissues.
Expression in disease
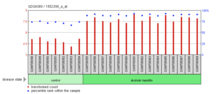
ERICH5 shows increased expression in Alcoholic Hepatitis.
Homology
Orthologs
True orthologs of ERICH5 have only been identified among mammalian species. The most distantly related mammalian ortholog is the Monodelphis domestica or the Gray Short Tailed Opossum.[20]
| Species Name | Common Names | MYA | % Similarity | % Identity | Accession # | Protein Length |
| Acinonyx jubatus | Cheetah | 96 | 71% | 65% | XP_014937513.1 | 358 |
| Cerathotherium | White Rhino | 96 | 71% | 66% | XP_004431343.2 | 358 |
| Trichechus manatus latirostris | Manatee | 105 | 68% | 59% | XP_004370879.3 | 342 |
| Monodelphis | Gray Short tailed opossum | 159 | 40% | 28% | XP_007488209.1 | 478 |
Distant orthologs
ERICH5 has distant orthologs among birds and reptiles. These distant orthologs contain only the third exon of ERICH5.[21]
| Species Name | Common Name | MYA | % Similarity | % Identity | Accession # | Protein Length |
| Gallus gallus | Chicken | 312 | 80% | 64% | XP_004940017.1 | 207 |
| Chysemys picta belli | Painted Turtle | 312 | 74% | 59% | XP_008166572.1 | 442 |
References
- GRCh38: Ensembl release 89: ENSG00000177459 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000044726 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Database, GeneCards Human Gene. "ERICH5 Gene - GeneCards | ERIC5 Protein | ERIC5 Antibody". www.genecards.org. Retrieved 2018-05-01.
- "ERICH5 glutamate rich 5 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2018-05-06.
- "NCBI Conserved Domain Search". www.ncbi.nlm.nih.gov. Retrieved 2018-05-07.
- "ELM - unknown". elm.eu.org. Retrieved 2018-05-07.
- "ELM - Detail for DEG_APCC_KENBOX_2". elm.eu.org. Retrieved 2018-05-07.
- "ELM - Detail for LIG_FHA_2". elm.eu.org. Retrieved 2018-05-07.
- "NetPhos 3.1 Server - prediction results". www.cbs.dtu.dk. Retrieved 2018-05-07.
- "NetOGlyc 4.0 Server - prediction results". www.cbs.dtu.dk. Retrieved 2018-05-07.
- "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interaction Motifs". sumosp.biocuckoo.org. Retrieved 2018-05-07.
- "Result for job ERICH5". raptorx.uchicago.edu. Retrieved 2018-05-07.
- "PSICQUIC View". EMBL-EBI.
- "PSORTII". PSORTII.
- Dezso, Z (2008). "A comprehensive functional analysis of tissue specificity of human gene expression". BMC Biology. 6: 49. doi:10.1186/1741-7007-6-49. PMC 2645369. PMID 19014478.
- "Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis".
- "Gene: ERICH5 (ENSG00000177459) - Comparative Genomics - Homo sapiens - Ensembl genome browser 92". useast.ensembl.org. Retrieved 2018-05-07.
- "BLAST: Basic Local Alignment Search Tool". blast.ncbi.nlm.nih.gov. Retrieved 2018-05-07.
- "BLAST: Basic Local Alignment Search Tool". blast.ncbi.nlm.nih.gov. Retrieved 2018-05-07.