Functional cloning
Functional cloning is a molecular cloning technique that relies on prior knowledge of the encoded protein’s sequence or function for gene identification.[1][2][3] In this assay, a genomic or cDNA library is screened to identify the genetic sequence of a protein of interest. Expression cDNA libraries may be screened with antibodies specific for the protein of interest or may rely on selection via the protein function.[1] Historically, the amino acid sequence of a protein was used to prepare degenerate oligonucleotides which were then probed against the library to identify the gene encoding the protein of interest.[2][3] Once candidate clones carrying the gene of interest are identified, they are sequenced and their identity is confirmed. This method of cloning allows researchers to screen entire genomes without prior knowledge of the location of the gene or the genetic sequence.[1]
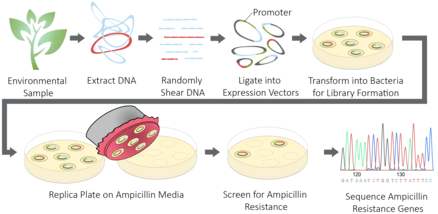
This technique can be used to identify genes that encode similar proteins from one organism to another.[4] Similarly, this technique can be paired with metagenomic libraries to identify novel genes and proteins that perform similar functions, such as the identification of novel antibiotics by screening for beta-lactamase activity or selecting for growth in the presence of penicillin.[5]
Experimental workflow

The workflow of a functional cloning experiment varies depending on the source of genetic material, the extent of prior knowledge of the protein or gene of interest and the ability to screen for the protein function. In general, a functional cloning experiment consists of four steps: 1) sample collection, 2) library preparation, 3) screening or selection and 4) sequencing.
Sample collection
Genetic material is collected from a particular cell type, organism or environmental sample relevant to the biological question. In functional cloning, mRNA is commonly isolated and cDNA is prepared from the isolated mRNA (RNA extraction).[6] In certain circumstances genomic DNA may be isolated, particularly when environmental samples are used as the source of genetic material.[1]
Library preparation
If the starting material is genomic DNA, the DNA is sheared to produce fragments of appropriate length for the vector of choice. The DNA fragments or cDNA are then treated with restriction endonucleases and ligated to a plasmid or chromosomal vectors. In the case of assays that screen for the protein or for its function, an expression vector is used to ensure that the gene product is expressed. The vector choice will depend on the origin of the DNA or cDNA to ensure proper expression and to ensure that the encoded gene will fall within the limits of the vector’s insert size.[7]
The choice of host is important to ensure that the codon usage will be similar to the donor organism. The host will also need to guarantee that the proper post-translational modifications and protein folding will occur to enable proper functioning of the expressed proteins.[7]
Screening or selection
The method of screening the prepared genomic or cDNA libraries for the gene of interest is highly variable depending on the experimental design and biological question. One method of screening is to probe colonies via Southern blotting with degenerate oligonucleotides prepared from the amino acid sequence of the query protein.[8] In expression libraries, the protein of interest can be identified by screening with an antibody specific for the query protein via Western blotting to identify colonies carrying the gene of interest. In other circumstances, a specific assay can be used to screen or select for the protein’s activity.[1] For example, genes conferring antibiotic resistance can be selected by growing the colonies of the library on media containing a specified antibiotic.[5] Another example is screening for enzymatic activity by incubating with a substrate that is catalyzed to a colorimetric compound that can easily be visualized.[9]
Sequencing
The final step of functional cloning is to sequence the DNA or cDNA from the clones that were successfully identified in the screen or selection step. The sequence can then be annotated and used for downstream applications, such as protein expression and purification for industrial applications.[10]
Advantages
The advantages of functional cloning include the ability to screen for novel genes with desired applications in organisms that cannot be cultured, particularly from bacterial or viral specimens.[1] Additionally, genes encoding proteins with related functions can be identified when there is low sequence similarity due to the ability to screen for the protein function alone. Functional cloning allows for gene identification without prior knowledge of the organism’s genome sequence or position of the gene within the genome.[1]
Limitations
As with other cloning techniques, vector and host choice affect the success of gene identification via functional cloning due to cloning bias. The vector must have an insert size that will accommodate the entire DNA sequence of the expressed protein. Additionally, in expression vectors the promoters and terminators must function within the chosen host organism. The host choice may affect transcription and translation due to differing codon usage, transcriptional and translational machinery or post-translational modifications within the host.[7][1]
Other limitations include the labour-intensive library preparation and potential screens which can be both expensive and time-consuming.[7]
Alternative approaches
Positional cloning
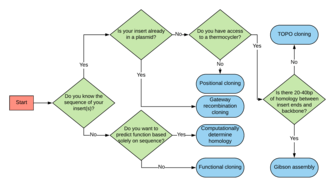
Positional cloning is another molecular cloning technique for identification of a gene of interest. This method uses exact chromosomal location instead of function to guide gene identification.[11] Because of this, this method focuses on all the genetic material at a chromosomal locus and makes no assumptions about function.[11] In model organisms such as mice or yeast, this method is used more frequently as the information about the position of a gene of interest can be obtained from the sequenced genome. However, this method becomes much more cumbersome when sequence information is not available. In this case, linkage analysis can also be used.[11] Functional cloning on the other hand is more readily used in organisms such as bacterial pathogens that are viable but nonculturable and where sequence data is not available but gene homology or protein function is still of interest.[11]
A way to differentiate between functional and positional cloning is to visualize genes as words. Functional cloning is like using a thesaurus to look up words and selecting for new words that have the same meanings (or functions).[12] Positional cloning is more like picking a specific page of a dictionary and then browsing only that page for any words of interest.[12]
Computationally determine homology
With the advent of sequencing technology becoming cheaper and cheaper, it is now more feasible to sequence an unknown genome and then computationally determine homology instead of screening.[13] This brings the added benefit of being able to screen for multiple genes of interest at the same time and reduces experimental time. It also allows one to avoid labour-intensive cloning procedures as well.[14] However, if this route is taken, there are other biases and hurdles one must consider. By using sequenced data, one is able to screen based on homology alone.[1] A function-based approach thus allows for discovery of novel enzymes whose functions would not have been predicted based on DNA sequence alone.[1] Therefore, while sequencing is less labour-intensive experimentally, it can also lead to missed genes of interest due to differing sequence homology in genes of related function.
Gibson assembly
Gibson assembly is a quick cloning method that uses three primary enzymes; 5' exonuclease, polymerase and ligase.[15] The exonuclease digests the 5' end of DNA fragments leaving a 3' overhang.[15] If there is significant homology (20-40 bp) on each end of the DNA insert, it can anneal with a complementary backbone.[15] Afterwards the polymerase can fill in the gaps while ligase fuses the nicks at the end.[15] This method greatly increases the rate of cloning and success rate of cloning into a vector backbone.[15] However, it requires the DNA fragment to have significant homology with the plasmid.[15] For this reason, knowledge of the sequence being cloned must be known beforehand. This is not a requirement with functional cloning.
TOPO cloning
TOPO Cloning is a cloning method that uses Taq polymerase.[16] This is because Taq leaves a single adenosine overhang on the 3’ end of PCR reaction products.[16] Utilizing this knowledge, backbones with a 5’ thymine overhang can be used for cloning purposes.[16] In this case knowledge of the fragment being cloned must be known to be able to make PCR primers for it and the number of TOPO Cloning compatible vectors is relatively small. However, it provides the advantage that reactions only take about 5 minutes to do.[16]
Gateway recombination cloning
Gateway recombination cloning is a cloning method in which a DNA fragment is moved from one plasmid backbone to another via a single homologous recombination event.[17] However, for this method to work, the DNA fragment of interest must be flanked by recombination sites.[17] While this method isn't strictly an alternative, it does allow the movement of DNA fragments from one plasmid to another quicker than creating a whole new genomic library. The reason this method may be used in conjunction with functional cloning is to put a library under a different promoter or on a backbone with a different selection marker.[17] This can come in handy if an individual wants to try functional cloning in a wide range of bacteria to try to combat the issue with codon bias.[17][7]
Applications
Determining homology in the environment
Metagenomics is one of the largest fields that commonly uses functional cloning. Metagenomics studies all the genetic material from a specific environmental sample, such as the gut microbiome or lake water.[1] Functional libraries are created that contain DNA fragments from the environment.[1] As the original bacterium that a DNA sequence originated from cannot be easily detected, creating metagenomic functional libraries possesses advantages. Less than 1% of all bacteria are easily cultured in the lab, leaving a large percentage of bacteria that cannot be grown.[18] By using functional libraries, the gene functions of unculturable bacteria can still be studied.[1] Furthermore, these uncultured microbes provide a source for the discovery of novel enzymes with biotechnological applications. Some novel proteins that have been discovered from marine environments include enzymes such as proteases, amylases, lipases, chitinases, deoxyribonucleases and phosphatases.[19]
Determining homology in a known species
There are situations in which it is imperative to determine if a gene homolog from one source is present in another organism. For example, identification of novel DNA polymerases for polymerase chain reaction (PCR) reactions which synthesize DNA molecules from deoxyribonucleotides.[20] While human polymerase optimally works at 37 °C (98.6 °F), DNA does not denature until 94–98 °C (201–208 °F).[20] This poses a problem as at these temperatures the human DNA polymerase would denature during the denaturation step of the PCR reaction resulting in a non-functioning polymerase protein and a failed PCR. To combat this a DNA polymerase from a thermophile, or bacteria that grows at high temperatures, could be used. An example is Taq polymerase which comes from the thermophilic bacterium Thermus aquaticus.[20] One could set up a functional cloning screen to find homologous polymerases that have the added advantage of being thermostable at high temperatures.
With this in mind, 3173 Polymerase, another polymerase enzyme, now commonly used in RT-PCR reactions was discovered using the above theory.[21] In RT-PCR reactions, two separate enzymes are commonly used. The first is a retroviral reverse transcriptase to convert RNA to cDNA.[21] The second is a thermostable DNA polymerase to amplify the target sequence.[21] 3173 Polymerase is able to perform both enzymatic functions resulting in a better option for RT-PCR.[21] The enzyme was discovered using functional cloning from a viral host originally found in Octopus hot springs (93 °C) in Yellowstone National Park.[21]
Human health applications
One of the ongoing challenges of treating bacterial infections is antibiotic resistance which commonly arises when patients do not take their full treatment of medication and hence allow bacteria to develop resistance to antibiotics over time.[22] To understand how to combat antibiotic resistance it is important to understand how the bacterial genome is evolving and changing in healthy individuals with no recent usage of antibiotics to provide a baseline.[22] Using a functional cloning-based technique, DNA isolated from human microflora were cloned into expression vectors in Escherichia coli.[22] Afterwards, antibiotics were applied as a screen.[22] If a plasmid contained a gene insert that provided antibiotic resistance the cell survived and was selected on the plate.[22] If the insert provided no resistance, the cell died and did not form a colony.[22] Based on selection of cell colonies that survived, a better picture of genetic factors contributing to antibiotic resistance were pieced together. Most of the resistance genes that were identified were previously unknown.[22] By using a functional cloning-based technique one is able to elucidate genes giving rise to antibiotic resistance to better understand treatment for bacterial infections.
References
- Lam, Kathy N.; Cheng, Jiujun; Engel, Katja; Neufeld, Josh D.; Charles, Trevor C. (2015). "Current and future resources for functional metagenomics". Frontiers in Microbiology. 6: 1196. doi:10.3389/fmicb.2015.01196. ISSN 1664-302X. PMC 4625089. PMID 26579102.
- Oxford dictionary of biochemistry and molecular biology. Cammack, Richard, Ph. D. (Rev. ed.). Oxford: Oxford University Press. 2006. ISBN 9780198529170. OCLC 65467611.CS1 maint: others (link)
- Schindler-Hoehn; Hoehn, Holger (2007-01-01). Fanconi Anemia: A Paradigmatic Disease for the Understanding of Cancer and Aging. Karger Medical and Scientific Publishers. ISBN 9783805582773.
- Skjøt, M.; Kauppinen, S.; Kofod, L.; Fuglsang, C.; Pauly, M.; Dalbøge, H.; Andersen, L. (2001-07-01). "Functional cloning of an endo-arabinanase from Aspergillus aculeatus and its heterologous expression in A. oryzae and tobacco". Molecular Genetics and Genomics. 265 (5): 913–921. doi:10.1007/s004380100489. ISSN 1617-4615.
- Allen, Heather K; Moe, Luke A; Rodbumrer, Jitsupang; Gaarder, Andra; Handelsman, Jo (2008-10-09). "Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil". The ISME Journal. 3 (2): 243–251. doi:10.1038/ismej.2008.86. ISSN 1751-7370. PMID 18843302.
- Shyjan, Anne M.; Heldin, Paraskevi; Butcher, Eugene C.; Yoshino, Tadashi; Briskin, Michael J. (1996-09-20). "Functional Cloning of the cDNA for a Human Hyaluronan Synthase". Journal of Biological Chemistry. 271 (38): 23395–23399. doi:10.1074/jbc.271.38.23395. ISSN 0021-9258. PMID 8798544.
- Lodish, Harvey; Berk, Arnold; Zipursky, S. Lawrence; Matsudaira, Paul; Baltimore, David; Darnell, James (2000). "Constructing DNA Libraries with λ Phage and Other Cloning Vectors". Cite journal requires
|journal=(help) - Mølhøj, Michael; Verma, Rajeev; Reiter, Wolf-Dieter (2004-07-01). "The Biosynthesis of d-Galacturonate in Plants. Functional Cloning and Characterization of a Membrane-Anchored UDP-d-Glucuronate 4-Epimerase from Arabidopsis". Plant Physiology. 135 (3): 1221–1230. doi:10.1104/pp.104.043745. ISSN 0032-0889. PMC 519042. PMID 15247385.
- Cheng, Jiujun; Romantsov, Tatyana; Engel, Katja; Doxey, Andrew C.; Rose, David R.; Neufeld, Josh D.; Charles, Trevor C. (2017-03-08). "Functional metagenomics reveals novel β-galactosidases not predictable from gene sequences". PLOS ONE. 12 (3): e0172545. Bibcode:2017PLoSO..1272545C. doi:10.1371/journal.pone.0172545. ISSN 1932-6203. PMC 5342196. PMID 28273103.
- Vester, Jan Kjølhede; Glaring, Mikkel Andreas; Stougaard, Peter (2014-05-20). "Discovery of novel enzymes with industrial potential from a cold and alkaline environment by a combination of functional metagenomics and culturing". Microbial Cell Factories. 13: 72. doi:10.1186/1475-2859-13-72. ISSN 1475-2859. PMC 4035831. PMID 24886068.
- Puliti, Aldamaria; Caridi, Gianluca; Ravazzolo, Roberto; Ghiggeri, Gian Marco (2007-12-01). "Teaching molecular genetics: chapter 4—positional cloning of genetic disorders". Pediatric Nephrology. 22 (12): 2023–2029. doi:10.1007/s00467-007-0548-5. ISSN 0931-041X. PMC 2908434. PMID 17661092.
- Rand, Elizabeth B. (1998-02-01). "The Genetic Basis of the Alagille Syndrome". Journal of Pediatric Gastroenterology & Nutrition. 26 (2): 234–236. doi:10.1097/00005176-199802000-00024. ISSN 0277-2116.
- Shendure, Jay; Balasubramanian, Shankar; Church, George M.; Gilbert, Walter; Rogers, Jane; Schloss, Jeffery A.; Waterston, Robert H. (2017). "DNA sequencing at 40: past, present and future". Nature. 550 (7676): 345–353. Bibcode:2017Natur.550..345S. doi:10.1038/nature24286. ISSN 1476-4687. PMID 29019985.
- "Microbial Whole-Genome Sequencing". www.illumina.com. Retrieved 2018-02-27.
- Wang, Jia-Wang; Wang, Amy; Li, Kunyu; Wang, Bangmei; Jin, Shunqian; Reiser, Michelle; Lockey, Richard F (2015). "CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning". BioTechniques. 58 (4): 161–70. doi:10.2144/000114261. PMID 25861928.
- Xu, Ruqiang; Qingshun, Li Quinn (2008-01-22). "Protocol: Streamline cloning of genes into binary vectors in Agrobacterium via the Gateway® TOPO vector system". Plant Methods. 4: 4. doi:10.1186/1746-4811-4-4. ISSN 1746-4811. PMC 2257932. PMID 18211693.
- Petersen, Lena K.; Stowers, R. Steven (2011-09-09). "A Gateway MultiSite Recombination Cloning Toolkit". PLOS ONE. 6 (9): e24531. Bibcode:2011PLoSO...624531P. doi:10.1371/journal.pone.0024531. ISSN 1932-6203. PMC 3170369. PMID 21931740.
- Amann, Rudolf (2000). "Who is out there? Microbial Aspects of Biodiversity". Systematic and Applied Microbiology. 23 (1): 1–8. doi:10.1016/s0723-2020(00)80039-9. PMID 10879972.
- Kennedy, Jonathan; Marchesi, Julian R.; Dobson, Alan DW (2008-08-21). "Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments". Microbial Cell Factories. 7: 27. doi:10.1186/1475-2859-7-27. ISSN 1475-2859. PMC 2538500. PMID 18717988.
- Garibyan, Lilit; Avashia, Nidhi (2013). "Polymerase Chain Reaction". Journal of Investigative Dermatology. 133 (3): 1–4. doi:10.1038/jid.2013.1. PMC 4102308. PMID 23399825.
- Moser, Michael J.; DiFrancesco, Robert A.; Gowda, Krishne; Klingele, Audrey J.; Sugar, Darby R.; Stocki, Stacy; Mead, David A.; Schoenfeld, Thomas W. (2012-06-04). "Thermostable DNA Polymerase from a Viral Metagenome Is a Potent RT-PCR Enzyme". PLOS ONE. 7 (6): e38371. Bibcode:2012PLoSO...738371M. doi:10.1371/journal.pone.0038371. ISSN 1932-6203. PMC 3366922. PMID 22675552.
- Sommer, Morten O. A.; Dantas, Gautam; Church, George M. (2009-08-28). "Functional Characterization of the Antibiotic Resistance Reservoir in the Human Microflora". Science. 325 (5944): 1128–1131. Bibcode:2009Sci...325.1128S. doi:10.1126/science.1176950. ISSN 0036-8075. PMC 4720503. PMID 19713526.
External links
- Functional Cloning, Sorting, and Expression Profiling of Nucleic Acid-Binding Proteins Genome Research.com
- A novel strategy for the functional cloning of enzymes using filamentous phage display: the case of nucleotidyl transferases Oxford Journals]
- Molecular genetics of Cohen syndrome Department of Medical Genetics, University of Helsinki