Gibson assembly
Gibson Assembly is a molecular cloning method which allows for the joining of multiple DNA fragments in a single, isothermal reaction. It is named after its creator, Daniel G. Gibson, who was the Chief Technology Officer and co-founder of Codex DNA.
Process
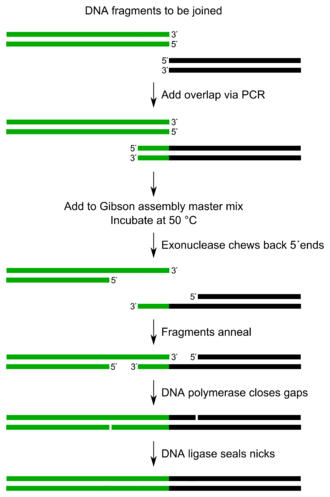
The entire Gibson Assembly reaction requires few components with minor manipulations.[1]
The method can simultaneously combine up to 15 DNA fragments based on sequence identity. It requires that the DNA fragments contain ~20-40 base pair overlap with adjacent DNA fragments. These DNA fragments are mixed with a cocktail of three enzymes, along with other buffer components.
The three required enzyme activities are: exonuclease, DNA polymerase, and DNA ligase.
- The exonuclease chews back DNA from the 5' end, thus not inhibiting polymerase activity and allowing the reaction to occur in one single process.[1] The resulting single-stranded regions on adjacent DNA fragments can anneal.
- The DNA polymerase incorporates nucleotides to fill in any gaps.
- The DNA ligase covalently joins the DNA of adjacent segments, thereby removing any nicks in the DNA.
The resulting product is different DNA fragments joined into one. Either linear or closed circular molecules can be assembled.
There are two approaches to Gibson Assembly. A one-step method and a two-step method. Both methods can be performed in a single reaction vessel. The Gibson Assembly 1-Step method allows for the assembly of up to 5 different fragments using a single step isothermal process. In this method, fragments and a master mix of enzymes are combined and the entire mixture is incubated at 50 °C for up to one hour. For the creation of more complex constructs with up to 15 fragments, or for constructs incorporating fragments from 100 bp to 10 kb, the Gibson Assembly two-step approach is used. The two-step reaction requires two separate additions of master mix; one for the exonuclease and annealing step and second for the DNA polymerase and ligation steps. For the two-step approach, different incubation temperatures are used to carry out the assembly process.
Advantages
This DNA assembly method has many advantages compared to conventional restriction enzyme/ligation cloning of recombinant DNA.
- No restriction digest of the DNA fragments after PCR is necessary. However, the backbone vector can be digested, or synthesized by PCR.
- It is cheaper and faster than conventional cloning schemes, as it requires fewer steps and fewer reagents.
- No restriction site scar remains between two DNA fragments, but the region between the double strands and hanging ends is slightly susceptible to mutation when DNA polymerase closes the gaps.
- Up to 5 DNA fragments can be combined simultaneously in a single-tube reaction using a one-step master mix of enzymes.
- Up to 15 fragments can be combined simultaneously using a two-step reaction. In the two step approach, the exonuclease and annealing steps are done first. This is followed by the addition of the DNA polymerase and ligase in a second step.
- The Gibson Assembly method can also be used for site directed mutagenesis to incorporate site-specific mutations such as insertions, deletions and point mutations
References
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009). "Enzymatic assembly of DNA molecules up to several hundred kilobases". Nature Methods. 6 (5): 343–345. doi:10.1038/nmeth.1318. PMID 19363495.
Further information
- A Guide to Gibson Assembly from the University of Cambridge, UK
- Gibson Assembly Primer Design Tool
- Gibson Assembly Site Directed Mutagenesis Primer Design Tool
- Chemical Transformation of Gibson Assembly Constructs
- Perkel, Jeffrey M. (January 2014). "Seamlessly rewriting the lab cloning manual". Tech News. BioTechniques. 56 (1): 12–14.
Gibson says he no longer even bothers with standard restriction enzyme-based cloning in his lab.
