Digital polymerase chain reaction
Digital polymerase chain reaction (digital PCR, DigitalPCR, dPCR, or dePCR) is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and traditional PCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users.[1] A "digital" measurement quantitatively and discretely measures a certain variable, whereas an “analog” measurement extrapolates certain measurements based on measured patterns. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences — such as copy number variants and point mutations — and it is routinely used for clonal amplification of samples for next-generation sequencing.
Principles
The polymerase chain reaction method is used to quantify nucleic acids by amplifying a nucleic acid molecule with the enzyme DNA polymerase.[2] Conventional PCR is based on the theory that amplification is exponential. Therefore, nucleic acids may be quantified by comparing the number of amplification cycles and amount of PCR end-product to those of a reference sample. However, many factors complicate this calculation, creating uncertainties and inaccuracies. These factors include the following: initial amplification cycles may not be exponential; PCR amplification eventually plateaus after an uncertain number of cycles; and low initial concentrations of target nucleic acid molecules may not amplify to detectable levels. However, the most significant limitation of PCR is that PCR amplification efficiency in a sample of interest may be different from that of reference samples. Since PCR is an exponential process, only twofold differences in amplification can be observed, greatly impacting the validity and precision of the results.

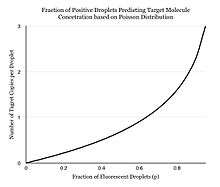
Instead of performing one reaction per well, dPCR involves partitioning the PCR solution into tens of thousands of nano-liter sized droplets, where a separate PCR reaction takes place in each one.[3][4] A PCR solution is made similarly to a TaqMan assay, which consists of template DNA (or RNA), fluorescence-quencher probes, primers, and a PCR master mix, which contains DNA polymerase, dNTPs, MgCl2, and reaction buffers at optimal concentrations. Several different methods can be used to partition samples, including microwell plates, capillaries, oil emulsion, and arrays of miniaturized chambers with nucleic acid binding surfaces.[5] The PCR solution is divided into smaller reactions and are then made to run PCR individually. After multiple PCR amplification cycles, the samples are checked for fluorescence with a binary readout of “0” or “1”. The fraction of fluorescing droplets is recorded.[4] The partitioning of the sample allows one to estimate the number of different molecules by assuming that the molecule population follows the Poisson distribution, thus accounting for the possibility of multiple target molecules inhabiting a single droplet. Using Poisson's law of small numbers, the distribution of target molecule within the sample can be accurately approximated allowing for a quantification of the target strand in the PCR product.[6] This model simply predicts that as the number of samples containing at least one target molecule increases, the probability of the samples containing more than one target molecule increases. In conventional PCR, the number of PCR amplification cycles is proportional to the starting copy number. Different from many peoples's belief that dPCR provides absolute quantification, digital PCR uses statistical power to provide relative quantification. For example, if Sample A, when assayed in 1 million partitions, gives one positive reaction, it does not mean that the Sample A has one starting molecule.
The benefits of dPCR include increased precision through massive sample partitioning, which ensures reliable measurements in the desired DNA sequence due to reproducibility.[4] Error rates are larger when detecting small-fold change differences with basic PCR, while error rates are smaller with dPCR due to the smaller-fold change differences that can be detected in DNA sequence. The technique itself reduces the use of a larger volume of reagent needed, which inevitably will lower experiment cost. Also, dPCR is highly quantitative as it does not rely on relative fluorescence of the solution to determine the amount of amplified target DNA.
Comparison between dPCR and Real-Time PCR (qPCR)
dPCR measures the actual number of molecules (target DNA) as each molecule is in one droplet, thus making it a discrete “digital” measurement. It provides absolute quantification because dPCR measures the positive fraction of samples, which is the number of droplets that are fluorescing due to proper amplification. This positive fraction accurately indicates the initial amount of template nucleic acid. Similarly, qPCR utilizes fluorescence; however, it measures the intensity of fluorescence at specific times (generally after every amplification cycle) to determine the relative amount of target molecule (DNA), but cannot specify the exact amount without constructing a standard curve using different amounts of a defined standard. It gives the threshold per cycle (CT) and the difference in CT is used to calculate the amount of initial nucleic acid. As such, qPCR is an analog measurement, which may not be as precise due to the extrapolation required to attain a measurement.[5][7]
dPCR measures the amount of DNA after amplification is complete and then determines the fraction of replicates. This is representative of an endpoint measurement as it requires the observation of the data after the experiment is completed. In contrast, qPCR records the relative fluorescence of the DNA at specific points during the amplification process, which requires stops in the experimental process. This “real-time” aspect of qPCR may theoretically affect results due to the stopping of the experiment. In practice, however, most qPCR thermal cyclers read each sample's fluorescence very quickly at the end of the annealing/extension step before proceeding to the next melting step, meaning this hypothetical concern is not actually relevant or applicable for the vast majority of researchers.
qPCR is unable to distinguish differences in gene expression or copy number variations that are smaller than twofold.[8] It is difficult to identify alleles with frequencies of less than 1% because highly abundant, common alleles would be matched with similar sequences. On the other hand, dPCR has been shown to detect differences of less than 30% in gene expression, distinguish between copy number variations that differ by only 1 copy, and identify alleles that occur at frequencies less than 0.1%.[9]
Applications
Digital PCR has many applications in basic research, clinical diagnostics and environmental testing. Its uses include pathogen detection and digestive health analysis;[10][11] liquid biopsy for cancer monitoring, organ transplant rejection monitoring and non-invasive prenatal testing for serious genetic abnormalities;[12][13][14][15][16][17][18][19] copy number variation analysis,[20][21][22] single gene expression analysis,[23] rare sequence detection,[19][24][25] gene expression profiling and single-cell analysis;[26][27][25][28][29][30][31] the detection of DNA contaminants in bioprocessing,[32] the validation of gene edits and detection of specific methylation changes in DNA as biomarkers of cancer.[33][34][35][36] dPCR is also frequently used as an orthogonal method to confirm rare mutations detected through next-generation sequencing (NGS) and to validate NGS libraries.[37][38][39]
Absolute quantification
dPCR enables the absolute and reproducible quantification of target nucleic acids at single-molecule resolution.[25][40][41][42] Unlike analogue quantitative PCR (qPCR), however, absolute quantification with dPCR does not require a standard curve).[40] dPCR also has a greater tolerance for inhibitor substances and PCR assays that amplify inefficiently as compared to qPCR.[43][44]
dPCR can quantify, for example, the presence of specific sequences from contaminating genetically modified organisms in foodstuffs,[45] viral load in the blood,[46] PBMCs,[47][48] serum samples,[49] chorionic villi tissues,[50][51] biomarkers of neurodegenerative disease in cerebral spinal fluid,[52] and fecal contamination in drinking water. [53]
Copy number variation
An alteration in copy number state with respect to a single-copy reference locus is referred to as a “copy number variation” (CNV) if it appears in germline cells, or a copy number alteration (CNA) if it appears in somatic cells.[54] A CNV or CNA could be due to a deletion or amplification of a locus with respect to the number of copies of the reference locus present in the cell, and together, they are major contributors to variability in the human genome.[55][56][57] They have been associated with cancers;[58][59][60] neurological,[61] psychiatric,[62][63] and autoimmune diseases;[64] and adverse drug reactions.[65] However, it is difficult to measure these allelic variations with high precision using other methods such as qPCR, thus making phenotypic and disease associations with altered CNV status challenging.[66][67]
The large number of “digitized,” endpoint measurements made possible by sample partitioning enables dPCR to resolve small differences in copy number with better accuracy and precision when compared to other methods such as SNP-based microarrays[68] or qPCR.[69][70] qPCR is limited in its ability to precisely quantify gene amplifications in several diseases, including Crohn’s disease, HIV-1 infection, and obesity.[71][67][70]
dPCR was designed to measure the concentration of a nucleic acid target in copies per unit volume of the sample. When operating in dilute reactions where less than ~10% of the partitions contain a desired target (referred to as “limiting dilution”), copy number can be estimated by comparing the number of fluorescent droplets arising from a target CNV with the number of fluorescent droplets arising from an invariant single-copy reference locus.[20] In fact, both at these lower target concentrations and at higher ones where multiple copies of the same target can co-localize to a single partition, Poisson statistics are used to correct for these multiple occupancies to give a more accurate value for each target’s concentration.[72][73]
Digital PCR has been used to uncover both germline and somatic variation in gene copy number between humans[74] and to study the link between amplification of HER2 (ERBB2) and breast cancer progression.[75][76][77][22]
Rare mutation and rare allele detection
Partitioning in digital PCR increases sensitivity and allows for detection of rare events, especially single nucleotide variants (SNVs), by isolating or greatly diminishing the target biomarker signal from potentially competing background.[7][5] These events can be organized into two classes: rare mutation detection and rare sequence detection.
Rare Mutation Detection
Rare mutation detection occurs when a biomarker exists within a background of a highly abundant counterpart that differs by only a single nucleotide variant (SNV). Digital PCR has been shown to be capable of detecting mutant DNA in the presence of a 200,000-fold excess of wild type background, which is 2,000 times more sensitive than achievable with conventional qPCR.[7]
Rare Sequence Detection
Digital PCR can detect rare sequences such as HIV DNA in patients with HIV,[19] and DNA from fecal bacteria in ocean and other water samples for assessing water quality.[78] dPCR can detect sequences as rare as 1 in every 1,250,000 cells.[19]
Liquid Biopsy
dPCR’s ability to detect rare mutations may be of particular benefit in the clinic through the use of the liquid biopsy, a generally noninvasive strategy for detecting and monitoring disease via bodily fluids.[12][79] Researchers have used liquid biopsy to monitor tumor load, treatment response and disease progression in cancer patients by measuring rare mutations in circulating tumor DNA (ctDNA) in a variety of biological fluids from patients including blood, urine and cerebrospinal fluid.[12][80][81]. Early detection of ctDNA (as in molecular relapse) may lead to earlier administration of an immunotherapy or a targeted therapy specific for the patient’s mutation signature, potentially improving chances of the treatment’s effectiveness rather than waiting for clinical relapse before altering treatment. Liquid biopsies can have turnaround times of a few days, compared to two to four weeks or longer for tissue-based tests.[82][83] This reduced time to results has been used by physicians to expedite treatments tailored to biopsy data.[82]
In 2016, a prospective trial using dPCR at the Dana-Farber Cancer Institute authenticated the clinical benefit of liquid biopsy as a predictive diagnostic tool for patients with non-small-cell lung cancer.[84] The application of liquid biopsy tests have also been studied in patients with breast,[85] colorectal,[86][87] gynecologic,[88] and bladder cancers[80][89] to monitor both the disease load and the tumor’s response to treatment.
Gene expression and RNA quantification
Gene expression and RNA quantification studies have benefited from the increased precision and absolute quantification of dPCR. RNA quantification can be accomplished via RT-PCR, wherein RNA is reverse-transcribed into cDNA in the partitioned reaction itself, and the number of RNA molecules originating from each transcript (or allelic transcript) is quantified via dPCR (ref).[26]
One can often achieve greater sensitivity and precision by using dPCR rather than qPCR to quantify RNA molecules in part because it does not require use of a standard curve for quantification.[90] dPCR is also more resilient to PCR inhibitors for the quantification of RNA than qPCR.[43][11]
dPCR can detect and quantify more individual target species per detection channel than qPCR by virtue of being able to distinguish targets based on their differential fluorescence amplitude or by the use of distinctive color combinations for their detection.[91] As an example of this, a 2-channel dPCR system has been used to detect in a single well the expression of four different splice variants of human telomerase reverse transcriptase, a protein that is more active in most tumor cells than in healthy cells.[92]
Alternative Uses for Partitioning
Using the dynamic partitioning capabilities employed in dPCR, improved NGS sequencing can be achieved by partitioning of complex PCR reactions prior to amplification to give more uniform amplification across many distinct amplicons for NGS analysis.[93][94] Additionally, the improved specificity of complex PCR amplification reactions in droplets has been shown to greatly reduce the number of iterations required to select for high affinity aptamers in the SELEX method.[95] Partitioning can also allow for more robust measurements of telomerase activity from cell lysates.[96][97] dPCR’s dynamic partitioning capabilities can also be used to partition thousands of nuclei or whole cells into individual droplets to facilitate library preparation for a single cell assay for transposase-accessible chromatin using sequencing (scATAC-seq).[98]
Droplet Digital PCR
Droplet Digital PCR (ddPCR) is a method of dPCR in which a 20 microliter sample reaction including assay primers and either Taqman probes or an intercalating dye, is divided into ~20,000 nanoliter-sized oil droplets through a water-oil emulsion technique, thermocycled to endpoint in a 96-well PCR plate, and fluorescence amplitude read for all droplets in each sample well in a droplet flow cytometer.[99]
History
dPCR rose out of an approach first published in 1988 by Cetus Corporation when researchers showed single β-globin molecules could be detected and amplified by PCR.[100][101] This was achieved by dividing the sample so some reactions contained the molecule and others did not. In 1990, Peter Simmonds and AJ Brown used this concept to quantify a molecule for the first time.[102] Alex Morley and Pamela Sykes formally established the method as a quantitative technique in 1992.[41]
In 1999, Bert Vogelstein and Kenneth Kinzler coined the term “digital PCR” and showed that the technique could be used to find rare cancer mutations.[103] However, dPCR was difficult to perform; it was labor intensive, required a lot of training to do properly, and was difficult to do in large quantities. [103] In 2003, Kinzler and Vogelstein continued to refine dPCR and created an improved method that they called BEAMing technology, an acronym for “beads, emulsion, amplification and magnetics.” The new protocol used emulsion to compartmentalize amplification reactions in a single tube. This change made it possible for scientists to scale the method to thousands of reactions in a single run.[104][105][106]
Companies developing commercial dPCR systems have integrated technologies like automated partitioning of samples, digital counting of nucleic acid targets, and increasing droplet count that can help the process be more efficient.[107][108][109] In recent years, scientists have developed and commercialized dPCR-based diagnostics for several conditions, including non-small cell lung cancer and Down’s Syndrome.[110][111] The first dPCR system for clinical use was CE-marked in 2017 and cleared by the US Food and Drug Administration in 2019, for diagnosing chronic myeloid leukemia.[112]
References
- Perkel J (May 2015). "Guiding our PCR experiments". BioTechniques. 58 (5): 217–21. doi:10.2144/000114283. PMID 25967899.
- "Polymerase Chain Reaction (PCR)". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Duewer, David L.; et al. (2018). "Evaluating droplet digital PCR for the quantification of human genomic DNA: converting copies per nanoliter to nanograms nuclear DNA per microliter". Analytical and Bioanalytical Chemistry. 410 (12): 2879–2887. doi:10.1007/s00216-018-0982-1. ISSN 1618-2642. PMC 5996397. PMID 29556737.
- Baker, Monya (2012). "Digital PCR hits its stride". Nature Methods. 9 (6): 541–544. doi:10.1038/nmeth.2027.
- Quan, Phenix-Lan; Sauzade, Martin; Brouzes, Eric (2018). "dPCR: A Technology Review". Sensors. 18 (4): 1271. doi:10.3390/s18041271. ISSN 1424-8220. PMC 5948698. PMID 29677144.
- Prediger E. "Digital PCR (dPCR)—What is it and why use it?". Integrated DNA Technologies.
- Pekin, Deniz; et al. (2011). "Quantitative and sensitive detection of rare mutations using droplet-based microfluidics". Lab on a Chip. 11 (13): 2156–66. doi:10.1039/c1lc20128j. ISSN 1473-0197. PMID 21594292.
- Ledger; et al. (2016). "Resolution of differences in gene expression using qPCR: a review of techniques". Journal of Biomolecular Technology. 42 (23): 230–245.
- Baker, Monya (2012-06-01). "Digital PCR hits its stride". Nature Methods. 9 (6): 541–544. doi:10.1038/nmeth.2027.
- Witte, Anna Kristina; et al. (2016). "Evaluation of the performance of quantitative detection of the Listeria monocytogenes prfA locus with droplet digital PCR". Analytical and Bioanalytical Chemistry. 408 (27): 7583–7593. doi:10.1007/s00216-016-9861-9. ISSN 1618-2642. PMC 5061835. PMID 27558101.
- Stauber, Jennifer; et al. (2016). "Droplet digital PCR quantifies host inflammatory transcripts in feces reliably and reproducibly". Cellular Immunology. 303: 43–49. doi:10.1016/j.cellimm.2016.03.007. ISSN 0008-8749. PMC 4863679. PMID 27063479.
- Skibo, Scott (23 Feb 2018). "Has Tumor Profiling Caught Up to Cancer?". Retrieved 23 July 2019.
- Hirsch, Fred (27 July 2018). "Guidelines highlight 'best practices' for liquid biopsy during treatment of non-small cell lung cancer". Retrieved 23 July 2019.
- Johnson, Madeleine (12 Jan 2018). "Bio-Rad Continues to Advance Digital PCR Tech, Liquid Biopsy Tests Into Commercial Clinical Market". Retrieved 23 July 2019.
- Oxnard, G. R.; et al. (2014). "Noninvasive Detection of Response and Resistance in EGFR-Mutant Lung Cancer Using Quantitative Next-Generation Genotyping of Cell-Free Plasma DNA". Clinical Cancer Research. 20 (6): 1698–1705. doi:10.1158/1078-0432.CCR-13-2482. ISSN 1078-0432. PMC 3959249. PMID 24429876.
- Schütz, E.; et al. (2017). "Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study". PLOS Medicine. 14 (4): e1002286. doi:10.1371/journal.pmed.1002286. PMC 5404754. PMID 28441386.
- Lee, S.Y.; Hwang, S.Y. (2015). "Application of digital polymerase chain reaction technology for noninvasive prenatal test". Journal of Genetic Medicine. 12 (2): 72–78. doi:10.5734/JGM.2015.12.2.72. ISSN 2383-8442.
- Gu, W.; et al. (2014). "Noninvasive prenatal diagnosis in a fetus at risk for methylmalonic acidemia". Genetics in Medicine. 16 (7): 564–567. doi:10.1038/gim.2013.194. PMC 4079742. PMID 24406457.
- Strain, M.; et al. (2013). "Highly Precise Measurement of HIV DNA by Droplet Digital PCR". PLOS ONE. 8 (4): e55943. Bibcode:2013PLoSO...855943S. doi:10.1371/journal.pone.0055943. PMC 3616050. PMID 23573183.
- Bell, Avery Davis; Usher, Christina L.; McCarroll, Steven A. (2018). "Analyzing Copy Number Variation with Droplet Digital PCR". Digital PCR. Methods in Molecular Biology. 1768. pp. 143–160. doi:10.1007/978-1-4939-7778-9_9. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717442.
- Shoda, Katsutoshi; et al. (2016). "Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer". Gastric Cancer. 20 (1): 126–135. doi:10.1007/s10120-016-0599-z. ISSN 1436-3291. PMID 26874951.
- Gevensleben, H.; et al. (2013). "Noninvasive Detection of HER2 Amplification with Plasma DNA Digital PCR". Clinical Cancer Research. 19 (12): 3276–3284. doi:10.1158/1078-0432.CCR-12-3768. ISSN 1078-0432. PMC 6485473. PMID 23637122.
- Torreggiani E, Rossini M, Bononi I, Pietrobon S, Mazzoni E, Iaquinta MR, Feo C, Rotondo JC, Rizzo P, Tognon M, Martini F (2019). "Protocol for the long-term culture of human primary keratinocytes from the normal colorectal mucosa". J Cell Physiol. 234 (7): 9895–9905. doi:10.1002/jcp.27490. PMID 30362540.
- Uchiyama, Yuri; et al. (2016). "Ultra–sensitive droplet digital PCR for detecting a low–prevalence somatic GNAQ mutation in Sturge–Weber syndrome". Scientific Reports. 6 (1): 22985. Bibcode:2016NatSR...622985U. doi:10.1038/srep22985. ISSN 2045-2322. PMC 4783707. PMID 26957145.
- Marusina, Kate (1 Oct 2017). "Positioning Digital PCR for Sharper Genomic Views". Retrieved 23 July 2019.
- Kamitaki, Nolan; et al. (2018). "Using Droplet Digital PCR to Analyze Allele-Specific RNA Expression". Digital PCR. Methods in Molecular Biology. 1768. pp. 401–422. doi:10.1007/978-1-4939-7778-9_23. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717456.
- Millier, Melanie J.; et al. (2017). "Digital-PCR for gene expression: impact from inherent tissue RNA degradation". Scientific Reports. 7 (1): 17235. Bibcode:2017NatSR...717235M. doi:10.1038/s41598-017-17619-0. ISSN 2045-2322. PMC 5722939. PMID 29222437.
- "Highly Sensitive Detection of Hepatitis B Using ddPCR". 12 Apr 2018. Retrieved 23 July 2019.
- Jang, Minjeong; et al. (2017). "Droplet-based digital PCR system for detection of single-cell level of foodborne pathogens". BioChip Journal. 11 (4): 329–337. doi:10.1007/s13206-017-1410-x. ISSN 2092-7843.
- Igarashi, Yuka; et al. (2017). "Single Cell-Based Vector Tracing in Patients with ADA-SCID Treated with Stem Cell Gene Therapy". Molecular Therapy - Methods & Clinical Development. 6: 8–16. doi:10.1016/j.omtm.2017.05.005. ISSN 2329-0501. PMC 5466583. PMID 28626778.
- Albayrak, Cem; et al. (2016). "Digital Quantification of Proteins and mRNA in Single Mammalian Cells". Molecular Cell. 61 (6): 914–24. doi:10.1016/j.molcel.2016.02.030. ISSN 1097-2765. PMID 26990994.
- Hussain, Musaddeq; et al. (2016). "A direct droplet digital PCR method for quantification of residual DNA in protein drugs produced in yeast cells". Journal of Pharmaceutical and Biomedical Analysis. 123: 128–131. doi:10.1016/j.jpba.2016.01.050. ISSN 0731-7085. PMID 26896631.
- Miyaoka, Yuichiro; et al. (2014). "Isolation of single-base genome-edited human iPS cells without antibiotic selection". Nature Methods. 11 (3): 291–293. doi:10.1038/nmeth.2840. PMC 4063274. PMID 24509632.
- Mock, Ulrike; et al. (2016). "Digital PCR to assess gene-editing frequencies (GEF-dPCR) mediated by designer nucleases". Nature Protocols. 11: 598–615. doi:10.1038/nmeth.2840. PMC 4063274. PMID 24509632.
- Nelson, C. E.; et al. (2015). "In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy". Science. 351 (6271): 403–407. doi:10.1126/science.aad5143. ISSN 0036-8075. PMC 4883596. PMID 26721684.
- Miyaoka, Yuichiro; et al. (2016). "Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing". Scientific Reports. 61: 23549. Bibcode:2016NatSR...623549M. doi:10.1038/srep23549. PMC 4814844. PMID 27030102.
- Guttery, D. S.; et al. (2015). "Noninvasive Detection of Activating Estrogen Receptor 1 (ESR1) Mutations in Estrogen Receptor-Positive Metastatic Breast Cancer". Clinical Chemistry. 61 (7): 974–982. doi:10.1373/clinchem.2015.238717. ISSN 0009-9147. PMID 25979954.
- Robin, Jérôme D.; et al. (2016). "Comparison of DNA Quantification Methods for Next Generation Sequencing". Scientific Reports. 6 (1): 24067. Bibcode:2016NatSR...624067R. doi:10.1038/srep24067. ISSN 2045-2322. PMC 4822169. PMID 27048884.
- Aigrain, Louise; et al. (2016). "Quantitation of next generation sequencing library preparation protocol efficiencies using droplet digital PCR assays - a systematic comparison of DNA library preparation kits for Illumina sequencing". BMC Genomics. 17 (1): 458. doi:10.1186/s12864-016-2757-4. ISSN 1471-2164. PMC 4906846. PMID 27297323.
- Brunetto, Giovanna S.; et al. (2014). "Digital droplet PCR (ddPCR) for the precise quantification of human T-lymphotropic virus 1 proviral loads in peripheral blood and cerebrospinal fluid of HAM/TSP patients and identification of viral mutations". Journal of NeuroVirology. 20 (4): 341–351. doi:10.1007/s13365-014-0249-3. ISSN 1355-0284. PMC 4085507. PMID 24781526.
- Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA (September 1992). "Quantitation of targets for PCR by use of limiting dilution". BioTechniques. 13 (3): 444–9. PMID 1389177.
- Vogelstein, B.; Kinzler, K. W. (1999). "Digital PCR". Proceedings of the National Academy of Sciences. 96 (16): 9236–9241. Bibcode:1999PNAS...96.9236V. doi:10.1073/pnas.96.16.9236. ISSN 0027-8424. PMC 17763. PMID 10430926.
- Rački, Nejc; et al. (2014). "Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples". Plant Methods. 10 (1): 42. doi:10.1186/s13007-014-0042-6. ISSN 1746-4811. PMC 4307183. PMID 25628753.
- Dingle, T. C.; et al. (2013). "Tolerance of Droplet-Digital PCR vs Real-Time Quantitative PCR to Inhibitory Substances". Clinical Chemistry. 59 (11): 1670–1672. doi:10.1373/clinchem.2013.211045. ISSN 0009-9147. PMC 4247175. PMID 24003063.
- Dobnik, David; et al. (2018). "Multiplex Droplet Digital PCR Protocols for Quantification of GM Maize Events". Digital PCR. Methods in Molecular Biology. 1768. pp. 69–98. doi:10.1007/978-1-4939-7778-9_5. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717438.
- Vellucci, Ashley; et al. (2018). "Using Droplet Digital PCR to Detect Coinfection of Human Herpesviruses 6A and 6B (HHV-6A and HHV-6B) in Clinical Samples". Digital PCR. Methods in Molecular Biology. 1768. pp. 99–109. doi:10.1007/978-1-4939-7778-9_6. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717439.
- Tagliapietra A, Rotondo JC, Bononi I, Mazzoni E, Magagnoli F, Maritati M (2019). "Droplet-digital PCR assay to detect Merkel cell polyomavirus sequences in chorionic villi from spontaneous abortion affected females". J Cell Physiol. 235 (3): 1888–1894. doi:10.1002/jcp.29213. PMID 31549405.
- Tagliapietra A, Rotondo JC, Bononi I, Mazzoni E, Magagnoli F, Maritati M, Contini C, Vesce F, Tognon M, Martini F (2019). "Footprints of BK and JC polyomaviruses in specimens from females affected by spontaneous abortion". Hum Reprod. 34 (3): 433–440. doi:10.1002/jcp.27490. PMID 30590693.
- Mazzoni E, Rotondo JC, Marracino L, Selvatici R, Bononi I, Torreggiani E, Touzé A, Martini F, Tognon MG (2017). "Detection of Merkel Cell Polyomavirus DNA in Serum Samples of Healthy Blood Donors". Front Oncol. 7: 433–440. doi:10.3389/fonc.2017.00294. PMC 5712532. PMID 29238698.
- Tagliapietra A, Rotondo JC, Bononi I, Mazzoni E, Magagnoli F, Maritati M (2019). "Droplet-digital PCR assay to detect Merkel cell polyomavirus sequences in chorionic villi from spontaneous abortion affected females". J Cell Physiol. 235 (3): 1888–1894. doi:10.1002/jcp.29213. PMID 31549405.
- Tagliapietra A, Rotondo JC, Bononi I, Mazzoni E, Magagnoli F, Maritati M, Contini C, Vesce F, Tognon M, Martini F (2019). "Footprints of BK and JC polyomaviruses in specimens from females affected by spontaneous abortion". Hum Reprod. 34 (3): 433–440. doi:10.1002/jcp.27490. PMID 30590693.
- Podlesniy, Petar; et al. (2018). "Biomarkers in Cerebrospinal Fluid: Analysis of Cell-Free Circulating Mitochondrial DNA by Digital PCR". Digital PCR. Methods in Molecular Biology. 1768. pp. 111–126. doi:10.1007/978-1-4939-7778-9_7. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717440.
- Cao, Yiping; et al. (2018). "Testing of General and Human-Associated Fecal Contamination in Waters". Digital PCR. Methods in Molecular Biology. 1768. pp. 127–140. doi:10.1007/978-1-4939-7778-9_8. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717441.
- Li, Wentian; et al. (2009). "Copy-number-variation and copy-number-alteration region detection by cumulative plots". BMC Bioinformatics. 10 (S1): S67. arXiv:0909.3129. Bibcode:2009arXiv0909.3129L. doi:10.1186/1471-2105-10-S1-S67. ISSN 1471-2105. PMC 2648736. PMID 19208171.
- Koren, Amnon; et al. (2014). "Genetic Variation in Human DNA Replication Timing". Cell. 159 (5): 1015–1026. doi:10.1016/j.cell.2014.10.025. ISSN 0092-8674. PMC 4359889. PMID 25416942.
- Sanders, Sean (16 Jul 2008). "CNVs vs SNPs: Understanding Human Structural Variation in Disease". Retrieved 24 July 2019.
- Marshall, Christian R; et al. (2016). "Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects". Nature Genetics. 49 (1): 27–35. doi:10.1038/ng.3725. ISSN 1061-4036. PMC 5737772. PMID 27869829.
- Shlien, Adam; Malkin, David (2009). "Copy number variations and cancer". Genome Medicine. 1 (6): 62. doi:10.1186/gm62. ISSN 1756-994X. PMC 2703871. PMID 19566914.
- Lauer, Stephanie; Gresham, David (2019). "An evolving view of copy number variants". Current Genetics. 65 (6): 1287–1295. doi:10.1007/s00294-019-00980-0. ISSN 0172-8083. PMID 31076843.
- "Copy Number Alteration Found to Be Associated with Cancer Mortality". 5 Sep 2018. Retrieved 24 July 2019.
- Gu, W.; Lupski, J.R. (2008). "CNV and nervous system diseases – what's new?". Cytogenetic and Genome Research. 123 (1–4): 54–64. doi:10.1159/000184692. ISSN 1424-8581. PMC 2920183. PMID 19287139.
- Thapar, Anita; Cooper, Miriam (2013). "Copy Number Variation: What Is It and What Has It Told Us About Child Psychiatric Disorders?". Journal of the American Academy of Child & Adolescent Psychiatry. 52 (8): 772–774. doi:10.1016/j.jaac.2013.05.013. ISSN 0890-8567. PMC 3919207. PMID 23880486.
- Sekar, Aswin; et al. (2016). "Schizophrenia risk from complex variation of complement component 4". Nature. 530 (7589): 177–183. Bibcode:2016Natur.530..177.. doi:10.1038/nature16549. ISSN 0890-8567. PMC 4752392. PMID 26814963.
- Yim, Seon-Hee; et al. (2015). "Clinical implications of copy number variations in autoimmune disorders". The Korean Journal of Internal Medicine. 30 (3): 294–304. doi:10.3904/kjim.2015.30.3.294. ISSN 1226-3303. PMC 4438283. PMID 25995659.
- He, Yijing; Hoskins, Janelle M.; McLeod, Howard L. (2011). "Copy number variants in pharmacogenetic genes". Trends in Molecular Medicine. 17 (5): 244–251. doi:10.1016/j.molmed.2011.01.007. ISSN 1471-4914. PMC 3092840. PMID 21388883.
- Gonzalez, E. (2005). "The Influence of CCL3L1 Gene-Containing Segmental Duplications on HIV-1/AIDS Susceptibility". Science. 307 (5714): 1434–1440. Bibcode:2005Sci...307.1434G. doi:10.1126/science.1101160. ISSN 0036-8075. PMID 15637236.
- Unutmaz, Derya; et al. (2010). "CCL3L1 Copy Number Variation and Susceptibility to HIV-1 Infection: A Meta-Analysis". PLOS ONE. 5 (12): e15778. Bibcode:2010PLoSO...515778L. doi:10.1371/journal.pone.0015778. ISSN 1932-6203. PMC 3012711. PMID 21209899.
- Dube, Simant; Qin, Jian; Ramakrishnan, Ramesh (2008). "Mathematical Analysis of Copy Number Variation in a DNA Sample Using Digital PCR on a Nanofluidic Device". PLOS ONE. 3 (8): e2876. Bibcode:2008PLoSO...3.2876D. doi:10.1371/journal.pone.0002876. ISSN 1932-6203. PMC 2483940. PMID 18682853.
- Hughesman, Curtis B.; et al. (2017). "Detection of clinically relevant copy number alterations in oral cancer progression using multiplexed droplet digital PCR". Scientific Reports. 7 (1): 11855. Bibcode:2017NatSR...711855H. doi:10.1038/s41598-017-11201-4. ISSN 2045-2322. PMC 5605662. PMID 28928368.
- Usher, Christina; et al. (2015). "Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity". Nature Genetics. 47 (8): 921–925. doi:10.1038/ng.3340. PMC 4712930. PMID 26098870.
- Aldhous, Marian C.; et al. (2010). "Measurement methods and accuracy in copy number variation: failure to replicate associations of beta-defensin copy number with Crohn's disease". Human Molecular Genetics. 19 (24): 4930–4938. doi:10.1093/hmg/ddq411. ISSN 1460-2083. PMC 2989891. PMID 20858604.
- Pinheiro, Leonardo; Emslie, Kerry R. (2018). "Basic Concepts and Validation of Digital PCR Measurements". Digital PCR. Methods in Molecular Biology. 1768. pp. 11–24. doi:10.1007/978-1-4939-7778-9_2. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717435.
- Quan, Phenix-Lan; Sauzade, Martin; Brouzes, Eric (2018). "dPCR: A Technology Review". Sensors. 18 (4): 1271. doi:10.3390/s18041271. ISSN 1424-8220. PMC 5948698. PMID 29677144.
- Handsaker, Robert E; et al. (2015). "Large multiallelic copy number variations in humans". Nature Genetics. 47 (3): 296–303. doi:10.1038/ng.3200. ISSN 1061-4036. PMC 4405206. PMID 25621458.
- Garcia-Murillas, Isaac; Turner, Nicholas C. (2018). "Assessing HER2 Amplification in Plasma cfDNA". Digital PCR. Methods in Molecular Biology. 1768. pp. 161–172. doi:10.1007/978-1-4939-7778-9_10. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717443.
- Christgen, Matthias; van Luttikhuizen; et al. (2016). "Precise ERBB2 copy number assessment in breast cancer by means of molecular inversion probe array analysis". Oncotarget. 7 (50): 82733–82740. doi:10.18632/oncotarget.12421. ISSN 1949-2553. PMC 5347728. PMID 27716627.
- Borley, A; et al. (2014). "Impact of HER2 copy number in IHC2+/FISH-amplified breast cancer on outcome of adjuvant trastuzumab treatment in a large UK cancer network". British Journal of Cancer. 110 (8): 2139–2143. doi:10.1038/bjc.2014.147. ISSN 0007-0920. PMC 3992505. PMID 24691421.
- Cao, Yiping; Raith, Meredith R.; Griffith, John F. (2015). "Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment". Water Research. 70: 337–349. doi:10.1016/j.watres.2014.12.008. ISSN 0043-1354. PMID 25543243.
- European Society for Medical Oncology (17 Nov 2017). "Study analyzes mutations in cerebrospinal fluid in lung cancer with brain metastases". Retrieved 24 July 2019.
- Petrone, Justin (8 Jun 2017). "Norwegian Team Plans to Debut Digital PCR-Based Urinary Bladder Cancer Test by Year End". Retrieved 24 July 2019.
- Hiemcke-Jiwa, Laura S.; et al. (2018). "The use of droplet digital PCR in liquid biopsies: A highly sensitive technique for MYD88 p.(L265P) detection in cerebrospinal fluid". Hematological Oncology. 36 (2): 429–435. doi:10.1002/hon.2489. PMID 29210102.
- Paxton, Anne (Oct 2017). "Revived hopes, fresh challenges with liquid biopsy". Retrieved 24 July 2019.
- Bhadra, Krish; Mellert, Hestia; Pestano, Gary (5 Jun 2017). "Adoption of Liquid Biopsy Tests for NSCLC". Retrieved 24 July 2019.
- Sacher, Adrian G.; Paweletz, Cloud; Dahlberg, Suzanne E. (2016). "Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer". JAMA Oncology. 2 (8): 1014–1022. doi:10.1001/jamaoncol.2016.0173. PMC 4982795. PMID 27055085.
- Olsson, Eleonor; et al. (2015). "Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease". EMBO Molecular Medicine. 7 (8): 1034–1047. doi:10.15252/emmm.201404913. ISSN 1757-4676. PMC 4551342. PMID 25987569.
- Carpinetti, Paola; et al. (2015). "The use of personalized biomarkers and liquid biopsies to monitor treatment response and disease recurrence in locally advanced rectal cancer after neoadjuvant chemoradiation". Oncotarget. 6 (35): 38360–71. doi:10.18632/oncotarget.5256. ISSN 1949-2553. PMC 4742005. PMID 26451609.
- Reinert, Thomas; et al. (2016). "Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery". Gut. 65 (4): 625–634. doi:10.1136/gutjnl-2014-308859. ISSN 0017-5749. PMID 25654990.
- Samimi, Goli; et al. (2015). "Personalized Circulating Tumor DNA Biomarkers Dynamically Predict Treatment Response and Survival In Gynecologic Cancers". PLOS ONE. 10 (12): e0145754. Bibcode:2015PLoSO..1045754P. doi:10.1371/journal.pone.0145754. ISSN 1932-6203. PMC 4696808. PMID 26717006.
- Dahmcke, Christina M.; et al. (2016). "A Prospective Blinded Evaluation of Urine-DNA Testing for Detection of Urothelial Bladder Carcinoma in Patients with Gross Hematuria". European Urology. 70 (6): 916–919. doi:10.1016/j.eururo.2016.06.035. ISSN 0302-2838. PMID 27417036.
- Taylor, Sean S.; et al. (2015). "Optimization of Droplet Digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: Clinical implications for quantification of Oseltamivir-resistant subpopulations". Journal of Virological Methods. 224: 58–66. doi:10.1016/j.jviromet.2015.08.014. PMID 26315318.
- Whale, Alexandra S.; Huggett, Jim F.; Tzonev, Svilen (2016). "Fundamentals of multiplexing with digital PCR". Biomolecular Detection and Quantification. 10: 15–23. doi:10.1016/j.bdq.2016.05.002. ISSN 2214-7535. PMC 5154634. PMID 27990345.
- Sun, Bing; Tao, Lian; Zheng, Yung-Ling (2014). "Simultaneous quantification of alternatively spliced transcripts in a single droplet digital PCR reaction". BioTechniques. 56 (6): 319–325. doi:10.2144/000114179. PMID 24924392.
- Valencia, C. Alexander; et al. (2012). "Assessment of Target Enrichment Platforms Using Massively Parallel Sequencing for the Mutation Detection for Congenital Muscular Dystrophy". The Journal of Molecular Diagnostics. 14 (3): 233–246. doi:10.1016/j.jmoldx.2012.01.009. ISSN 1525-1578. PMC 3349841. PMID 22426012.
- Brusgaard, Klaus; et al. (2015). "What Is the Best NGS Enrichment Method for the Molecular Diagnosis of Monogenic Diabetes and Obesity?". PLOS ONE. 10 (11): e0143373. Bibcode:2015PLoSO..1043373P. doi:10.1371/journal.pone.0143373. ISSN 1932-6203. PMC 4657897. PMID 26599467.
- Ouellet, Eric; et al. (2015). "Hi-Fi SELEX: A high-fidelity digital-PCR based therapeutic aptamer discovery platform". Biotechnology and Bioengineering. 112 (8): 1506–1522. doi:10.1002/bit.25581. ISSN 0006-3592. PMID 25727321.
- Ludlow, Andrew T.; et al. (2018). "ddTRAP: A Method for Sensitive and Precise Quantification of Telomerase Activity". Digital PCR. Methods in Molecular Biology. 1768. pp. 513–529. doi:10.1007/978-1-4939-7778-9_29. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMC 6046637. PMID 29717462.
- Sayed, Mohammed E.; Slusher, Aaron L.; Ludlow, Andrew T. (2019). "Droplet Digital TRAP (ddTRAP): Adaptation of the Telomere Repeat Amplification Protocol to Droplet Digital Polymerase Chain Reaction". Journal of Visualized Experiments (147). doi:10.3791/59550. ISSN 1940-087X. PMID 31107456.
- Stein, Richard A. (1 Jul 2019). "Single-Cell Sequencing Sifts through Multiple Omics". Retrieved 1 August 2019.
- Wood-Bouwens, Christina M.; Ji, Hanlee P. (2018). "Single Color Multiplexed ddPCR Copy Number Measurements and Single Nucleotide Variant Genotyping". Digital PCR. Methods in Molecular Biology. 1768. pp. 323–333. doi:10.1007/978-1-4939-7778-9_18. ISBN 978-1-4939-7776-5. ISSN 1064-3745. PMID 29717451.
- Erlich, H. A.; Mullis, K. B.; Horn, G. T.; Higuchi, R.; Scharf, S. J.; Stoffel, S.; Gelfand, D. H.; Saiki, R. K. (29 January 1988). "Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase". Science. 239 (4839): 487–491. Bibcode:1988Sci...239..487S. doi:10.1126/science.239.4839.487. ISSN 0036-8075. PMID 2448875.
- Morley, Alexander A. (1 September 2014). "Digital PCR: A brief history". Biomolecular Detection and Quantification. 1 (1): 1–2. doi:10.1016/j.bdq.2014.06.001. ISSN 2214-7535. PMC 5129430. PMID 27920991.
- Rutsaert, Sofie; Bosman, Kobus; Trypsteen, Wim; Nijhuis, Monique; Vandekerckhove, Linos (30 January 2018). "Digital PCR as a tool to measure HIV persistence". Retrovirology. 15 (1): 16. doi:10.1186/s12977-018-0399-0. ISSN 1742-4690. PMC 5789538. PMID 29378600.
- Perkel, Jeff (11 April 2014). "The digital PCR revolution". Retrieved 22 July 2019.
- Pohl G, Shih I (January 2004). "Principle and applications of digital PCR". Expert Review of Molecular Diagnostics. 4 (1): 41–7. doi:10.1586/14737159.4.1.41. PMID 14711348.
- Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B (July 2003). "Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations". Proceedings of the National Academy of Sciences of the United States of America. 100 (15): 8817–22. Bibcode:2003PNAS..100.8817D. doi:10.1073/pnas.1133470100. PMC 166396. PMID 12857956.
- Diehl F, Li M, Kinzler, KW, Vogelstein B, Dressman D (2006). "BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions". Nature Methods. 3 (7): 551–559. doi:10.1038/nmeth898. PMID 16791214.
- Butkus, Ben (8 July 2010). "Digital PCR Space Heating Up as Life Science Tool Vendors Begin Staking Claims". Retrieved 22 July 2019.
- Ramakrishnan R, Qin J, Jones RC, Weaver LS (2013). "Integrated Fluidic Circuits (IFCs) for digital PCR". Microfluidic Diagnostics. Methods in Molecular Biology. 949. pp. 423–31. doi:10.1007/978-1-62703-134-9_27. ISBN 978-1-62703-133-2. PMID 23329458.
- Butkus, Ben (29 Mar 2012). "RainDance Launches Digital PCR Platform; Claims Sensitivity, Operating Cost Superiority". Retrieved 22 July 2019.
- "'Liquid biopsy' blood test detects genetic mutations in common form of lung cancer". 7 Apr 2016. Retrieved 22 July 2019.
- "Korea's BioCore First to Commercialize NIPT Based on Digital PCR". 2 Mar 2018. Retrieved 22 July 2019.
- "Bio-Rad Gets First CE Mark on Clinical ddPCR Test". 5 Dec 2017. Retrieved 22 Jul 2019.