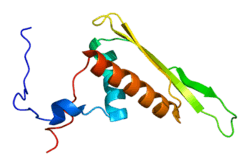RNA Helicase A
ATP-dependent RNA helicase A (RHA; also known as DHX9, LKP, and NDHI) is an enzyme that in humans is encoded by the DHX9 gene.[5][6][7]
Function
DEAD/DEAH box helicases are proteins, and are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein with RNA helicase activity. It may participate in melting of DNA:RNA hybrids, such as those that occur during transcription, and may play a role in X-linked gene expression. It contains 2 copies of a double-stranded RNA-binding domain, a DEXH core domain and an RGG box. The RNA-binding domains and RGG box influence and regulate RNA helicase activity.[7]The DHX9 gene is located on the long arm q of chromosome 1.
Interactions
DHX9 has been shown to interact with:
References
- GRCh38: Ensembl release 89: ENSG00000135829 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000042699 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Lee CG, Hurwitz J (Aug 1993). "Human RNA helicase A is homologous to the maleless protein of Drosophila". The Journal of Biological Chemistry. 268 (22): 16822–30. PMID 8344961.
- Zhang S, Grosse F (Apr 1997). "Domain structure of human nuclear DNA helicase II (RNA helicase A)". The Journal of Biological Chemistry. 272 (17): 11487–94. doi:10.1074/jbc.272.17.11487. PMID 9111062.
- "Entrez Gene: DHX9 DEAH (Asp-Glu-Ala-His) box polypeptide 9".
- Yang JP, Tang H, Reddy TR, Wong-Staal F (Aug 2001). "Mapping the functional domains of HAP95, a protein that binds RNA helicase A and activates the constitutive transport element of type D retroviruses". The Journal of Biological Chemistry. 276 (33): 30694–700. doi:10.1074/jbc.M102809200. PMID 11402034.
- Westberg C, Yang JP, Tang H, Reddy TR, Wong-Staal F (Jul 2000). "A novel shuttle protein binds to RNA helicase A and activates the retroviral constitutive transport element". The Journal of Biological Chemistry. 275 (28): 21396–401. doi:10.1074/jbc.M909887199. PMID 10748171.
- Schlegel BP, Starita LM, Parvin JD (Feb 2003). "Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells". Oncogene. 22 (7): 983–91. doi:10.1038/sj.onc.1206195. PMID 12592385.
- Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD (Jul 1998). "BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A". Nature Genetics. 19 (3): 254–6. doi:10.1038/930. PMID 9662397.
- Wilson BJ, Giguère V (November 2007). "Identification of novel pathway partners of p68 and p72 RNA helicases through Oncomine meta-analysis". BMC Genomics. 8: 419. doi:10.1186/1471-2164-8-419. PMC 3225811. PMID 18005418.
- Reddy TR, Tang H, Xu W, Wong-Staal F (Jul 2000). "Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA". Oncogene. 19 (32): 3570–5. doi:10.1038/sj.onc.1203676. PMID 10951562.
- Fujita H, Fujii R, Aratani S, Amano T, Fukamizu A, Nakajima T (Apr 2003). "Antithetic effects of MBD2a on gene regulation". Molecular and Cellular Biology. 23 (8): 2645–57. doi:10.1128/mcb.23.8.2645-2657.2003. PMC 152551. PMID 12665568.
- Tang H, Wong-Staal F (Oct 2000). "Specific interaction between RNA helicase A and Tap, two cellular proteins that bind to the constitutive transport element of type D retrovirus". The Journal of Biological Chemistry. 275 (42): 32694–700. doi:10.1074/jbc.M003933200. PMID 10924507.
- Smith WA, Schurter BT, Wong-Staal F, David M (May 2004). "Arginine methylation of RNA helicase a determines its subcellular localization". The Journal of Biological Chemistry. 279 (22): 22795–8. doi:10.1074/jbc.C300512200. PMID 15084609.
- Tetsuka T, Uranishi H, Sanda T, Asamitsu K, Yang JP, Wong-Staal F, Okamoto T (Sep 2004). "RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator". European Journal of Biochemistry / FEBS. 271 (18): 3741–51. doi:10.1111/j.1432-1033.2004.04314.x. PMID 15355351.
- Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G (Jan 2001). "A functional interaction between the survival motor neuron complex and RNA polymerase II". The Journal of Cell Biology. 152 (1): 75–85. doi:10.1083/jcb.152.1.75. PMC 2193649. PMID 11149922.
Further reading
- Lee CG, Hurwitz J (Mar 1992). "A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3' to 5' direction". The Journal of Biological Chemistry. 267 (7): 4398–407. PMID 1537828.
- Lee CG, Zamore PD, Green MR, Hurwitz J (Jun 1993). "RNA annealing activity is intrinsically associated with U2AF". The Journal of Biological Chemistry. 268 (18): 13472–8. PMID 7685763.
- Abdelhaleem MM, Hameed S, Klassen D, Greenberg AH (Mar 1996). "Leukophysin: an RNA helicase A-related molecule identified in cytotoxic T cell granules and vesicles". Journal of Immunology. 156 (6): 2026–35. PMID 8690889.
- Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M (Sep 1997). "RNA helicase A mediates association of CBP with RNA polymerase II". Cell. 90 (6): 1107–12. doi:10.1016/S0092-8674(00)80376-1. PMID 9323138.
- Loor G, Zhang SJ, Zhang P, Toomey NL, Lee MY (Dec 1997). "Identification of DNA replication and cell cycle proteins that interact with PCNA". Nucleic Acids Research. 25 (24): 5041–6. doi:10.1093/nar/25.24.5041. PMC 147130. PMID 9396813.
- Lee CG, Eki T, Okumura K, da Costa Soares V, Hurwitz J (Feb 1998). "Molecular analysis of the cDNA and genomic DNA encoding mouse RNA helicase A". Genomics. 47 (3): 365–71. doi:10.1006/geno.1997.5139. PMID 9480750.
- Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD (Jul 1998). "BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A". Nature Genetics. 19 (3): 254–6. doi:10.1038/930. PMID 9662397.
- Lee CG, da Costa Soares V, Newberger C, Manova K, Lacy E, Hurwitz J (Nov 1998). "RNA helicase A is essential for normal gastrulation". Proceedings of the National Academy of Sciences of the United States of America. 95 (23): 13709–13. doi:10.1073/pnas.95.23.13709. PMC 24884. PMID 9811865.
- Li J, Tang H, Mullen TM, Westberg C, Reddy TR, Rose DW, Wong-Staal F (Jan 1999). "A role for RNA helicase A in post-transcriptional regulation of HIV type 1". Proceedings of the National Academy of Sciences of the United States of America. 96 (2): 709–14. doi:10.1073/pnas.96.2.709. PMC 15201. PMID 9892698.
- Tang H, McDonald D, Middlesworth T, Hope TJ, Wong-Staal F (May 1999). "The carboxyl terminus of RNA helicase A contains a bidirectional nuclear transport domain". Molecular and Cellular Biology. 19 (5): 3540–50. doi:10.1128/mcb.19.5.3540. PMC 84146. PMID 10207077.
- Westberg C, Yang JP, Tang H, Reddy TR, Wong-Staal F (Jul 2000). "A novel shuttle protein binds to RNA helicase A and activates the retroviral constitutive transport element". The Journal of Biological Chemistry. 275 (28): 21396–401. doi:10.1074/jbc.M909887199. PMID 10748171.
- Tang H, Wong-Staal F (Oct 2000). "Specific interaction between RNA helicase A and Tap, two cellular proteins that bind to the constitutive transport element of type D retrovirus". The Journal of Biological Chemistry. 275 (42): 32694–700. doi:10.1074/jbc.M003933200. PMID 10924507.
- Lee CG, Eki T, Okumura K, Nogami M, Soares Vda C, Murakami Y, Hanaoka F, Hurwitz J (Jan 1999). "The human RNA helicase A (DDX9) gene maps to the prostate cancer susceptibility locus at chromosome band 1q25 and its pseudogene (DDX9P) to 13q22, respectively". Somatic Cell and Molecular Genetics. 25 (1): 33–9. doi:10.1023/B:SCAM.0000007138.44216.3a. PMID 10925702.
- Reddy TR, Tang H, Xu W, Wong-Staal F (Jul 2000). "Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA". Oncogene. 19 (32): 3570–5. doi:10.1038/sj.onc.1203676. PMID 10951562.
- Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G (Jan 2001). "A functional interaction between the survival motor neuron complex and RNA polymerase II". The Journal of Cell Biology. 152 (1): 75–85. doi:10.1083/jcb.152.1.75. PMC 2193649. PMID 11149922.
- Takasaki Y, Kogure T, Takeuchi K, Kaneda K, Yano T, Hirokawa K, Hirose S, Shirai T, Hashimoto H (Apr 2001). "Reactivity of anti-proliferating cell nuclear antigen (PCNA) murine monoclonal antibodies and human autoantibodies to the PCNA multiprotein complexes involved in cell proliferation". Journal of Immunology. 166 (7): 4780–7. doi:10.4049/jimmunol.166.7.4780. PMID 11254741.
- Yang JP, Tang H, Reddy TR, Wong-Staal F (Aug 2001). "Mapping the functional domains of HAP95, a protein that binds RNA helicase A and activates the constitutive transport element of type D retroviruses". The Journal of Biological Chemistry. 276 (33): 30694–700. doi:10.1074/jbc.M102809200. PMID 11402034.
- Aratani S, Fujii R, Oishi T, Fujita H, Amano T, Ohshima T, Hagiwara M, Fukamizu A, Nakajima T (Jul 2001). "Dual roles of RNA helicase A in CREB-dependent transcription". Molecular and Cellular Biology. 21 (14): 4460–9. doi:10.1128/MCB.21.14.4460-4469.2001. PMC 87106. PMID 11416126.