Curacin A
Curacin A is a hybrid polyketide synthase (PKS)/nonribosomal peptide synthase (NRPS) derived natural product produced isolated from the cyanobacterium Lyngbya majuscula.[1] Curacin A belongs to a family of natural products including jamaicamide, mupirocin, and pederin that have an unusual terminal alkene. Additionally, Curacin A contains a notable thiazoline ring and a unique cyclopropyl moiety, which is essential to the compound's biological activity.[1][2] Curacin A has been characterized as potent antiproliferative cytotoxic compound with notable anticancer activity for several cancer lines including renal, colon, and breast cancer.[2][3] Curacin A has been shown to interact with colchicine binding sites on tubulin, which inhibits microtubule polymerization, an essential process for cell division and proliferation.[1][4]
 | |
| Identifiers | |
|---|---|
| |
| CAS Number | |
| PubChem CID | |
| ChemSpider | |
| KEGG | |
| ChEBI | |
| ChEMBL | |
| Chemical and physical data | |
| Formula | C23H35NOS |
| Molar mass | 373.60 g·mol−1 |
| 3D model (JSmol) | |
| |
| |
| (verify) | |
Biosynthesis
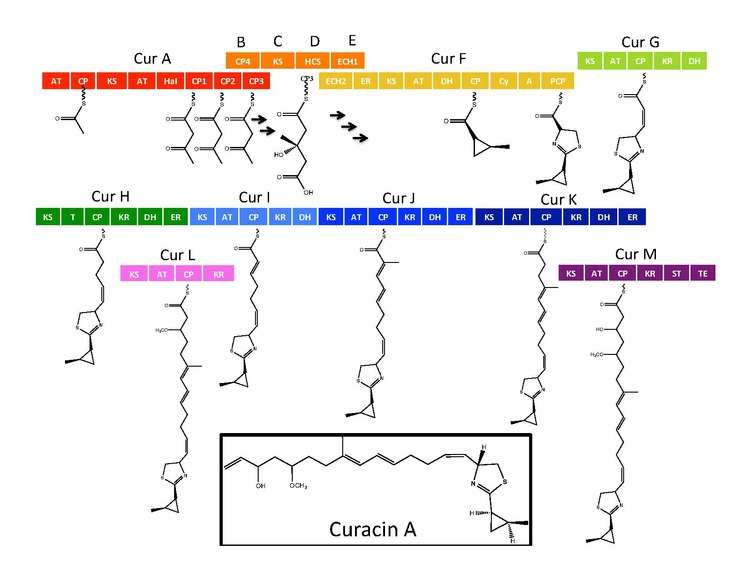
The synthetic enzymes for Curacin A are found in a gene cluster with 14 open reading frames (ORFs) with the nomenclature CurA through CurN.[1] Analysis of the pathway demonstrated the presence of one NRPS/PKS hybrid module located on CurF, one HMG-CoA synthase cassette located on CurD, and seven monomodular PKS modules.[1] CurA contains a unique GCN5-related N-acetyltransferase (GNAT) loading domain and an associated acyl carrier protein (ACP).[2] The loading module tethers an acetyl group to the ACP that then condenses with one of three tandem ACPs present in the adjacent module of CurA.[1][2][5] An hydroxymethylglutaryl-CoA synthase cassette (mevalonate pathway) catlyzes the formation of hydroxymethylglutaryl acid by the addition of an malonyl-CoA unit to the terminal ketide of the aceto-acetyl-ACP moiety of ACP1,ACP2, or ACP3.[5] subsequent enzymes, including a unique heme independent halogenase (HaI) catalyze the formation of a cyclopropyl ring.[1][5][6] A cysteine specific NRPS module located on CurF follows after cyclopropyl ring formation, and due to the activity of a cyclizing condensation domain, forms a thiazole ring attached to the cylcopropyl moiety from previous reactions in the pathway.[1][5][6] Seven standalone PKS modules follow to extend the growing polyketide chain with S-adenosyl methionine (SAM) dependent methylations occurring at positions 10 and 13.[1] A rare offloading strategy involving a sulfotransferase is employed by the final curacin synthase module. The sulfotransferase sulfates the hydroxyl group of carbon 15, which activates the molecule for decarboxylation and terminal alkene formation.[7]
Cyclopropyl ring formation
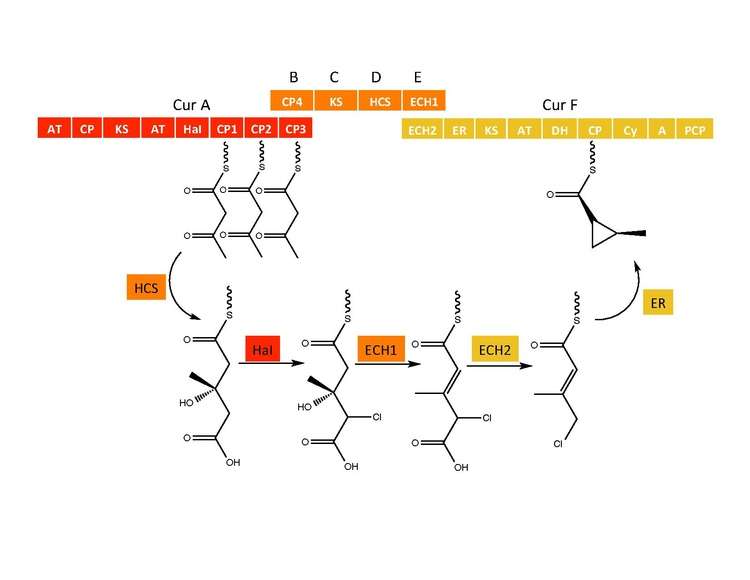
The CurB (ACP), CurC (ketosynthase), and CurD (HMG-CoA reductase) are responsible for the formation of (S)HMG-ACP3.[6] HaI, from the CurA gene, is a unique non-heme halogenase that goes through a purported Fe(IV)=O intermediate to add a chlorine atom onto an unactivated carbon atom.[6] After chlorination, ECH1 acting as a dehydratates HMG-ACP3 to 3-methylgultaconyl-ACP3 and ECH2 performs the required decarboxylation.[6] Finally,an unusual ER catalyzed cyclization reaction, purported to go through a substitution like mechanism, forms the cyclopropane ring.[6] The added chlorine atom assists in the decarboxylation step and likely serves as the leaving group during cyclopropane ring formation.[6]
References
- Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, et al. (August 2004). "Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula". Journal of Natural Products. 67 (8): 1356–67. doi:10.1021/np0499261. PMID 15332855.
- Gu L, Geders TW, Wang B, Gerwick WH, Håkansson K, Smith JL, Sherman DH (November 2007). "GNAT-like strategy for polyketide chain initiation". Science. 318 (5852): 970–4. Bibcode:2007Sci...318..970G. doi:10.1126/science.1148790. PMID 17991863.
- Verdier-Pinard P, Lai JY, Yoo HD, Yu J, Marquez B, Nagle DG, et al. (January 1998). "Structure-activity analysis of the interaction of curacin A, the potent colchicine site antimitotic agent, with tubulin and effects of analogs on the growth of MCF-7 breast cancer cells". Molecular Pharmacology. 53 (1): 62–76. doi:10.1124/mol.53.1.62. PMID 9443933.
- Blokhin AV, Yoo HD, Geralds RS, Nagle DG, Gerwick WH, Hamel E (September 1995). "Characterization of the interaction of the marine cyanobacterial natural product curacin A with the colchicine site of tubulin and initial structure-activity studies with analogues". Molecular Pharmacology. 48 (3): 523–31. PMID 7565634.
- Gu L, Eisman EB, Dutta S, Franzmann TM, Walter S, Gerwick WH, et al. (March 2011). "Tandem acyl carrier proteins in the curacin biosynthetic pathway promote consecutive multienzyme reactions with a synergistic effect". Angewandte Chemie. 50 (12): 2795–8. doi:10.1002/anie.201005280. PMC 3081611. PMID 21387490.
- Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, et al. (June 2009). "Metamorphic enzyme assembly in polyketide diversification". Nature. 459 (7247): 731–5. Bibcode:2009Natur.459..731G. doi:10.1038/nature07870. PMC 2918389. PMID 19494914.
- McCarthy JG, Eisman EB, Kulkarni S, Gerwick L, Gerwick WH, Wipf P, et al. (December 2012). "Structural basis of functional group activation by sulfotransferases in complex metabolic pathways". ACS Chemical Biology. 7 (12): 1994–2003. doi:10.1021/cb300385m. PMC 3528841. PMID 22991895.