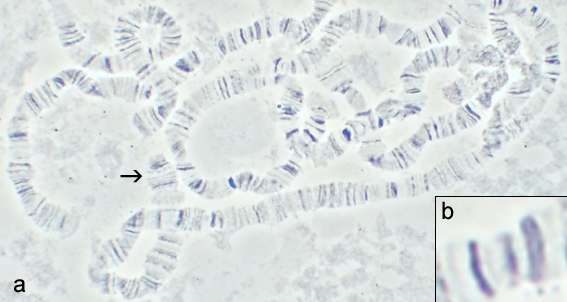
Chromomere
A chromomere, also known as an idiomere, is one of the serially aligned beads or granules of a eukaryotic chromosome, resulting from local coiling of a continuous DNA thread.[1] Chromeres are regions of chromatin that have been compacted through localized contraction. In areas of chromatin with the absence of transcription, condensing of DNA and protein complexes will result in the formation of chromomeres.[2] It is visible on a chromosome during the prophase of meiosis and mitosis. Giant banded (Polytene) chromosomes resulting from the replication of the chromosomes and the synapsis of homologs without cell division is a process called endomitosis. These chromosomes consist of more than 1000 copies of the same chromatid that are aligned and produce alternating dark and light bands when stained. The dark bands are the chromomere.
It is unknown when chromomeres first appear on the chromosome. Chromomeres can be observed best when chromosomes are highly condensed.[2] The chromomeres are present during leptotene phase of prophase I during meiosis. During zygotene phase of prophase I, the chromomeres of homologs align with each other to form homologous rough pairing (homology searching). These chromomeres helps provide a unique identity for each homologous pairs. They appear as dense granules during leptotene stage
There are more than 2000 chromomeres on 20 chromosomes of maize.
Physical properties
Chromomeres are organized in a discontinuous linear pattern along the condensed chromosomes (pachytene chromosomes) during the prophase stage of meiosis.[2] The linear pattern of chromomeres is linked to the arrangement of genes along the chromosome.[2][3] Chromomeres contain genes and sometimes clusters of genes within their structure.[3] Aggregates of chromomeres are known as chromonemata.[4]
Cohesive proteins SMC3 and hRAD21(plays a role in sister chromatid cohesion) are found within chromomeres at high concentrations, and maintain the proper structure of chromomeres. The protein XCAP-D2 is also present at high concentrations within the chromomere, and acts as a condensin component. High concentrations of tandem repeats of the heterochromatin protein HP1β builds up within the chromomere. In regions where loops attach to chromomeres, there is hyperacetylation of histone 4. The extra acetylation loosens chromatin from a condensed form, making it more accessible to proteins involved in transcription.[2]
Functions
Chromomeres are known as the structural subunit of a chromosome. The arrangement of chromomere structure can aid in control of gene expression.[3]
Maps of chromomeres can be made for use in genetic and evolutionary studies. Chromomeric maps can be used to locate the exact position of genes on a chromosome. Chromomeric maps can be used to analyze chromosome aberrations, and find correlations between the aberrations and their effects on genes near breakpoints.[3]
Lampbrush chromosomes and chromomeres
Chromomeres display different properties and behaviours when associated with lampbrush chromosomes. Although found all across the lampbrush chromosome, they are not organized in a clear pattern along as they are in normal pachytene chromosomes of meiosis.[2][5] The two sister chromatids of a lampbrush chromosome separate fully, forming lateral loops that extend from chromomeres, and act as transcription complexes. The lateral loops are areas where the chromosomes are transcriptionally active.[2][5] The loops define where the chromomere will be positioned. The chromomere is an organization of regions of non-transcribed chromatin. These regions of chromatin that have not been transcribed are located at the ends of the loops that were formed by the sister chromatids of a lampbrush chromosome.[2] Each chromomere can have up to several pairs of loops from lampbrush chromosomes originating from it, as well as micro-loops that cannot be detected with a light microscope.[6]
Chromomeres of lampbrush chromosomes act as structural units, and are not considered to be genetic units participating in transcription. The arrangement of a chromosome into chromomeric units happens almost simultaneously with the start of transcription.[2] The loop structures extending from chromomeres are maintained by a high level of transcriptional activity and structural proteins of the chromosome. Chromatin within the chromomere are held in position by a variety of histone modifications and epigenetic markers.[5]
Methyl-CpG-binding protein 2 (MeCP2) is a protein that binds to methylated DNA. MeCP2 has been found to associate most strongly with transcriptionally inactive chromomere domains. MeCP2 binding patterns to chromomere domains are proportional to the density of chromomeric DNA found on a chromosome. The binding of MeCP2 to chromomeric regions represents regions of chromatin that are highly dynamic on the lampbrush chromosome.[6]
References
- Mosby (2016-04-29). Mosby's Medical Dictionary - E-Book. Elsevier Health Sciences. pp. 362–. ISBN 978-0-323-41426-5.
- Macgregor, Herbert (September 7, 2012). "Chromomeres revisited". Chromosome Research. 20 (8): 911–924. doi:10.1007/s10577-012-9310-3. PMID 22956230. Retrieved 2015-09-24.
- Judd, Burke (September 1998). "Genes and chromomeres: A puzzle in three dimensions". Genetics. 150 (1): 1–9. PMC 1460313. PMID 9725825. Retrieved 2015-10-16.
- Sheval EV, Prusov AN, Kireev II, Fais D, Polyakov VY (2002). "Organization of higher-level chromatin structures (chromomere, chromonema and chromatin block) examined using visible light-induced chromatin photo-stabilization". Cell Biol. Int. 26 (7): 579–91. doi:10.1006/cbir.2002.0879. PMID 12127937.
- Gaginskaya E, Kulikova T, Krasikova A (2009). "Avian lampbrush chromosomes: a powerful tool for exploration of genome expression". Cytogenet. Genome Res. 124 (3–4): 251–67. doi:10.1159/000218130. PMID 19556778.
- Morgan, Garry (December 2012). "Association of modified cytosines and the methylated DNA-binding protein MeCP2 with distinctive structural domains of lampbrush chromatin". Chromosome Research. 20 (8): 925–42. doi:10.1007/s10577-012-9324-x. PMC 3565088. PMID 23149574.
External links
- "Preparation and analysis of spermatocyte meiotic pachytene bivalents of pigs for gene mapping" - Nature
- "Physical mapping of DNA repetitive sequences to mitotic and meiotic chromosomes of Brassica oleracea var. alboglabra by fluorescence in situ hybridization" - Nature
- "Chromonema and chromomere" - Springerlink