Brian (software)
Brian is an open source Python package for developing simulations of networks of spiking neurons.
| Developer(s) | Romain Brette, Dan Goodman, Marcel Stimberg |
|---|---|
| Stable release | 2.3
/ January 6, 2020 |
| Repository | |
| Written in | Python |
| Operating system | Cross-platform |
| Type | Neural network software |
| License | CeCILL |
| Website | http://www.briansimulator.org |
Details
Brian is aimed at researchers developing models based on networks of spiking neurons. The general design is aimed at maximising flexibility, simplicity and users' development time.[1] Users specify neuron models by giving their differential equations in standard mathematical form as strings, create groups of neurons and connect them via synapses. This is in contrast to the approach taken by many neural simulators in which users select from a predefined set of neuron models.
Brian is written in Python. Computationally, it is based around the concept of code generation: users specify the model in Python but behind the scenes Brian generates, compiles and runs code in one of several languages (including Python, Cython and C++). In addition there is a "standalone" mode in which Brian generates an entire C++ source code tree with no dependency on Brian, allowing models to be run on platforms where Python is not available.
Example
The following code defines, runs and plots a randomly connected network of leaky integrate and fire neurons with exponential inhibitory and excitatory currents.
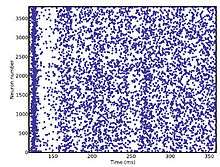
from brian2 import *
eqs = """
dv/dt = (ge+gi-(v+49*mV))/(20*ms) : volt
dge/dt = -ge/(5*ms) : volt
dgi/dt = -gi/(10*ms) : volt
"""
P = NeuronGroup(4000, eqs, threshold="v>-50*mV", reset="v=-60*mV")
P.v = -60 * mV
Pe = P[:3200]
Pi = P[3200:]
Ce = Synapses(Pe, P, on_pre="ge+=1.62*mV")
Ce.connect(p=0.02)
Ci = Synapses(Pi, P, on_pre="gi-=9*mV")
Ci.connect(p=0.02)
M = SpikeMonitor(P)
run(1 * second)
plot(M.t / ms, M.i, ".")
show()
Comparison to other simulators
Brian is primarily, although not solely, aimed at single compartment neuron models. Simulators focused on multi-compartmental models include Neuron, GENESIS, and its derivatives.
The focus of Brian is on flexibility and ease of use, and only supports simulations running on a single machine. The NEST simulator includes facilities for distributing simulations across a cluster.[2]
Footnotes
- Goodman and Brette 2009
- "NEST Initiative".
References
- Goodman, D. and Brette, R. (2008). "Brian: a simulator for spiking neural networks in Python", Front. Neuroinform. 2:5. doi:10.3389/neuro.11.005.2008 PMID 19115011
- Goodman, D.F.M. and Brette, R. (2009). "The Brian simulator", Front. Neurosci. doi:10.3389/neuro.01.026.2009