Backcrossing
Backcrossing is a crossing of a hybrid with one of its parents or an individual genetically similar to its parent, in order to achieve offspring with a genetic identity which is closer to that of the parent. It is used in horticulture, animal breeding and in production of gene knockout organisms.
Backcrossed hybrids are sometimes described with acronym "BC", for example, an F1 hybrid crossed with one of its parents (or a genetically similar individual) can be termed a BC1 hybrid, and a further cross of the BC1 hybrid to the same parent (or a genetically similar individual) produces a BC2 hybrid.[1]
Plants
Advantages
- If the recurrent parent is an elite genotype, at the end of the backcrossing programme an elite genotype is recovered.
- As there is no "new" recombination, the elite combination is not lost.
Disadvantages
- Works poorly for quantitative traits
- Is more restricted for recessive traits
- In practice, sections of genome from the non-recurrent parents are often still present and can have unwanted traits associated with them
- For very wide crosses, limited recombination may maintain thousands of ‘alien’ genes within the elite cultivar
- Many backcrosses are required to produce a new cultivar which can take many years
Natural backcrossings
York radiate groundsel (Senecio eboracensis) is a naturally occurring hybrid species of Oxford ragwort (Senecio squalidus) and common groundsel (Senecio vulgaris). It is thought to have arisen from a backcrossing of the F1 hybrid with S. vulgaris.[2]
Again, the pure tall (TT) and pure dwarf (tt) pea plants when crossed in the parental generation, they produce all heterozygote (Tt) tall pea plants in the first filial generation. The cross between first filial heterozygote tall (Tt) pea plant and pure tall (TT) or pure dwarf (tt) pea plant of the parental generation is also an example for the back-crossing between two plants. In this case, the filial generation formed after the back cross may have a phenotype ratio of 1:1 if the cross is made with recessive parent or else all offspring may be having phenotype of dominant trait if back cross is with parent having dominant trait. The former of these traits is also called a test cross.
Artificially recombinant lines
In plants, inbred backcross lines (IBLs) refers to lines (i.e. populations) of plants derived from the repeated backcrossing of a line with artificially recombinant DNA with the wild type, operating some kind of selection that can be phenotypical or through a molecular marker (for the production of introgression lines).
Animals
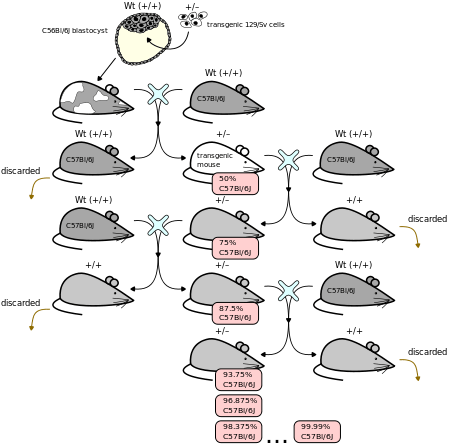
Backcrossing may be deliberately employed in animals to transfer a desirable trait in an animal of inferior genetic background to an animal of preferable genetic background. In gene knockout experiments in particular, where the knockout is performed on easily cultured stem cell lines, but is required in an animal with a different genetic background, the knockout animal is backcrossed against the animal of the required genetic background. As the figure shows, each time that the mouse with the desired trait (in this case the lack of a gene (i.e. a knockout), indicated by the presence of a positive selectable marker) is crossed with a mouse of a constant genetic background, the average percentage of the genetic material of the offspring that is derived from that constant background increases. The result, after sufficient reiterations, is an animal with the desired trait in the desired genetic background, with the percentage of genetic material from the original stem cells reduced to a minimum (in the order of 0.01%).[3]
Due to the nature of meiosis, in which chromosomes derived from each parent are randomly shuffled and assigned to each nascent gamete, the percentage of genetic material deriving from either cell-line will vary between offspring of a single crossing but will have an expected value. The genotype of each member of offspring may be assessed to choose not only an individual that carries the desired genetic trait, but also the minimum percentage of genetic material from the original stem cell line.[4]
A consomic strain is an inbred strain with one of its chromosomes replaced by the homologous chromosome of another inbred strain via a series of marker-assisted backcrosses.[5]
See also
References
Notes
- Schweitzer, J.A.; Martinsen, G.D.; Whitham, T.G. (2002). "Cottonwood hybrids gain fitness traits of both parents: a mechanism for their long-term persistence?". American Journal of Botany. 89 (6): 981–990. doi:10.3732/ajb.89.6.981. PMID 21665697.
- Abbot, R.J.; Lowe, A.J. (2003). "A new British species, Senecio eboracensis (Asteraceae), another hybrid derivative of S. vulgaris L. and S. squalidae L" (PDF). Watsonia. 24: 375–388. Archived from the original (PDF) on 2007-09-27. Retrieved 2007-07-15.
- "Embryonic Stem Cell". Archived from the original on 2006-09-09. Retrieved 2008-01-01.
- Frisch M, Melchinger AE (2005). "Selection theory for marker-assisted backcrossing". Genetics. 170 (2): 909–17. doi:10.1534/genetics.104.035451. PMC 1450430. PMID 15802512.
- The Jackson Laboratory > Consomic strains Archived 2011-06-24 at the Wayback Machine Last Modified: June 11, 2010
Bibliography
External links
- The Plant Breeding and Genomics Community of Practice on eXtension - education and training materials for plant breeders and allied professionals