Atg1
AuTophaGy related 1 (Atg1) is a 101.7kDa serine/threonine kinase in S.cerevisiae, encoded by the gene ATG1.[1] It is essential for the initial building of the autophagosome and Cvt vesicles. In a non-kinase role it is - through complex formation with Atg13 and Atg17 - directly controlled by the TOR kinase, a sensor for nutrient availability.
| Serine/threonine-protein kinase ATG1 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | ATG1 | ||||||
| Alt. symbols | APG1; AUT3; CVT10 | ||||||
| Entrez | 852695 | ||||||
| RefSeq (mRNA) | NM_001181045 | ||||||
| RefSeq (Prot) | NP_011335 | ||||||
| UniProt | P53104 | ||||||
| Other data | |||||||
| EC number | 2.7.11.1 | ||||||
| Chromosome | VII: 0.16 - 0.16 Mb | ||||||
| |||||||
Introduction
Atg1 can associate with a number of other proteins of the Atg family to form a complex that functions in autophagosome or Cvt vesicle formation. The initiation of autophagy involves the building of the pre-autophagosomal structure (PAS). Most Atg proteins accumulate at the PAS and generate either Cvt vesicles under normal growing conditions or autophagosomes under starvation.[2] To date, there are 31 ATG genes, which can be classified into several different groups according to their functions at the different steps of the pathway. 17 of these genes only work in the Cvt pathway.
Structure
The Atg1 gene lies on chromosome VII of S. cerevisiae. The encoded protein with a mass of 101.7 kDa has a length of 897 amino acids and includes a protein serine/threonin kinase domain of 302 amino acids at its N-terminus. At the C-terminus, there is a 7 amino acid long region that is required for Cvt trafficking. The protein is also post-translationally modified through phosphorylation of at least 9 serine residues [3] Until now, no crystal structure has been made of Atg1.
Function
Atg1 has two distinct functions in yeast (for higher eukaryotes see below): the kinase-independent recruitment of downstream Atg proteins (i.e. PAS organization) and a kinase-dependent function in autophagosome formation likely mediated by the phosphorylation of downstream substrates.
Interaction partners
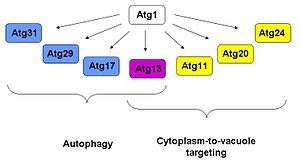
Atg1 has been shown to interact with at least six other Atg proteins, namely Atg 29, 31, 11, 20 and 24. Of all these, Atg13 has been shown to have roles in both autophagy and Cvt functions; Atg17, 29 and 31 have only functions in autophagy,[4][5][6] while Atg11, 20 and 24 only take part in the Cvt pathway.[7][8] Based on yeast two-hybrid data and affinity isolation, Atg1 is found to be in a complex with Atg13 and Atg17.[9] The observation that Atg17 interacts with Atg13 in the absence of Atg1 but not vice versa suggests that Atg13 mediates the interaction between Atg1 and Atg17.
Regulation
The autophagy machinery is activated upon several stimuli, like nutrient starvation, infection, repair mechanism or in programmed cell death. The role of Atg1 and its regulation is best studied under nutrient starvation and the corresponding arrest of growth. A key enzyme in the signalling pathway of nutrient availability is TOR, of which two isoforms exist in yeast(Tor1 and Tor2). These proteins form two distinct complexes, termed TORC1 and TORC2 of which TORC1 is highly sensitive to cellular nutrient conditions. Under nutrient rich conditions, TORC1 is active and phosphorylates Atg13 at multiple sites, thereby inhibiting a complex formation with Atg1. This leads to a decrease in Atg1 kinase activity and decreased autophagy. Upon starvation, Atg13 is rapidly dephosphorylated and forms a complex with Atg1, thereby activating it, which leads to the subsequent assembly of the PAS through recruitment of other Atg proteins.
In addition to TORC1, protein kinase A (PKA) inhibits autophagy through the phosphorylation of Atg1 and Atg13. PKA phosphorylates Atg1 at two distinct serine residues; these modifications were shown to be necessary for Atg1 to properly dissociate from the PAS.[10] The downstream substrate of Atg1 kinase hasn't been described yet, and it is still a matter of debate as to whether Atg1 primarily acts on autophagy through its kinase activity or through a structural role during autophagic complex formation. It is possible that the kinase activity of Atg1 is critical to the magnitude of autophagy but not its initiation. At least, large-scale screens led to a candidate list of possible Atg1 substrates, including Atg8 and Atg18.[11] In conclusion, Atg1 first has a structural or scaffolding function during the initial steps of PAS set up, which then is followed by a kinase-dependent phase, that contains protein dynamics at the PAS.[12]
Homologues
There is much evidence indicating that Atg1 homologues from other, multicellular organisms are required for autophagy as well but Recent work however also showed that there are differences and additional functions compared to the yeast model.
Caenorhabditis elegans
The corresponding homologue to Atg1 in C. elegans is unc-51 (uncoordinated-51). Unc-51 also functions in proper axonal guidance and in neuronal development.[13]
Drosophila melanogaster
The Atg1 homologue in D. melanogaster is also important in neural development [14] and neuronal trafficking. Additionally there is a feed back mechanism to TOR, that can inhibit TOR function, which actually lies upstream of Atg1.[15] Atg1 and Atg13 are always in one complex in 'D.melanogaster and vertebrates. In D.melanogaster, Atg13 gets phosphorylated in starvation, what is exactly the opposite as in the yeast model.
Vertebrates
There are until now five potential Atg1 orthologues in vertebrates. ULK1 and ULK2 (unc-51-like kinase) have been reported to have an additional function in neuronal development, e.g. outgrowth regulation of mouse neurons.[16] ULK1 and 2 also show a negative feedback regulation to mTOR.
References
- "ATG1 Summary". YeastGenome.org. Retrieved 4 January 2012.
- Mizushima N (January 2010). "The role of the Atg1/ULK1 complex in autophagy regulation". Curr Opin Cell Biol. 22 (2): 132–139. doi:10.1016/j.ceb.2009.12.004. PMID 20056399.
- "UniProt P53104 (ATG1_YEAST)". UniProtKB. March 2, 2010. Retrieved March 17, 2010.
- Kawamata T, Kamada Y, Suzuki K, et al. (December 2005). "Characterization of a novel autophagy-specific gene, ATG29". Biochem. Biophys. Res. Commun. 338 (4): 1884–9. doi:10.1016/j.bbrc.2005.10.163. PMID 16289106.
- Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y (May 2007). "Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae". Biochem. Biophys. Res. Commun. 356 (2): 405–10. doi:10.1016/j.bbrc.2007.02.150. PMID 17362880.
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (May 2008). "Organization of the pre-autophagosomal structure responsible for autophagosome formation". Mol. Biol. Cell. 19 (5): 2039–50. doi:10.1091/mbc.E07-10-1048. PMC 2366851. PMID 18287526.
- Kim J, Kamada Y, Stromhaug PE, et al. (April 2001). "Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole". J. Cell Biol. 153 (2): 381–96. doi:10.1083/jcb.153.2.381. PMC 2169458. PMID 11309418.
- Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ (August 2002). "Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy". J. Biol. Chem. 277 (33): 30198–207. doi:10.1074/jbc.M204736200. PMC 2754692. PMID 12048214.
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y (May 2005). "Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy". Mol. Biol. Cell. 16 (5): 2544–53. doi:10.1091/mbc.E04-08-0669. PMC 1087256. PMID 15743910.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (September 2000). "Tor-mediated induction of autophagy via an Apg1 protein kinase complex". J. Cell Biol. 150 (6): 1507–13. doi:10.1083/jcb.150.6.1507. PMC 2150712. PMID 10995454.
- Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK (September 2005). "An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase". Proc. Natl. Acad. Sci. U.S.A. 102 (39): 13933–8. doi:10.1073/pnas.0501046102. PMC 1236527. PMID 16172400.
- Ptacek J, Devgan G, Michaud G, et al. (December 2005). "Global analysis of protein phosphorylation in yeast" (PDF). Nature. 438 (7068): 679–84. doi:10.1038/nature04187. PMID 16319894.
- Chan EY, Tooze SA (August 2009). "Evolution of Atg1 function and regulation". Autophagy. 5 (6): 758–65. doi:10.4161/auto.8709. PMID 19411825.
- Ogura K, Wicky C, Magnenat L, et al. (October 1994). "Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase". Genes Dev. 8 (20): 2389–400. doi:10.1101/gad.8.20.2389. PMID 7958904.
- Toda H, Mochizuki H, Flores R, et al. (December 2008). "UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly". Genes Dev. 22 (23): 3292–307. doi:10.1101/gad.1734608. PMC 2600757. PMID 19056884.
- Scott RC, Juhász G, Neufeld TP (January 2007). "Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death". Curr. Biol. 17 (1): 1–11. doi:10.1016/j.cub.2006.10.053. PMC 1865528. PMID 17208179.
- Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME (December 1999). "A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons". Neuron. 24 (4): 833–46. doi:10.1016/S0896-6273(00)81031-4. PMID 10624947.