Alpha sheet
Alpha sheet (also known as alpha pleated sheet or polar pleated sheet) is an atypical secondary structure in proteins, first proposed by Linus Pauling and Robert Corey in 1951.[1][2][3] The hydrogen bonding pattern in an alpha sheet is similar to that of a beta sheet, but the orientation of the carbonyl and amino groups in the peptide bond units is distinctive; in a single strand, all the carbonyl groups are oriented in the same direction on one side of the pleat, and all the amino groups are oriented in the same direction on the opposite side of the sheet. Thus the alpha sheet accumulates an inherent separation of electrostatic charge, with one edge of the sheet exposing negatively charged carbonyl groups and the opposite edge exposing positively charged amino groups. Unlike the alpha helix and beta sheet, the alpha sheet configuration does not require all component amino acid residues to lie within a single region of dihedral angles; instead, the alpha sheet contains residues of alternating dihedrals in the traditional right-handed (αR) and left-handed (αL) helical regions of Ramachandran space. Although the alpha sheet is only rarely observed in natural protein structures, it has been speculated to play a role in amyloid disease[4] and it was found to be a stable form for amyloidogenic proteins in molecular dynamics simulations.[5][6] Alpha sheets have also been observed in X-ray crystallography structures of designed peptides.[4]
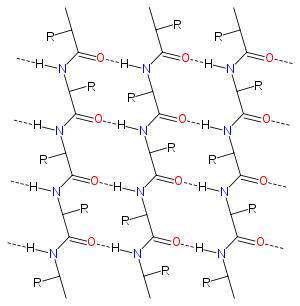 Diagram of the hydrogen bonding patterns in the alpha sheet structure. Oxygen atoms are shown in red and nitrogen in blue; dotted lines represent hydrogen bonds. R groups represent the amino acid side chains. |
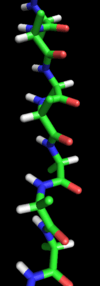 A stick representation of a peptide chain in an alpha-sheet configuration. |
Experimental evidence
When Pauling and Corey first proposed the alpha sheet, they suggested that it agreed well with fiber diffraction results from beta-keratin fibers.[2] However, since the alpha sheet did not appear to be energetically favorable, they argued that beta sheets would occur more commonly among normal proteins,[3] and subsequent demonstration that beta-keratin is made of beta sheets consigned the alpha sheet proposal to obscurity. However the alpha strand conformation is observed in isolated instances in native state proteins as solved by X-ray crystallography or protein NMR, although an extended alpha sheet is not identified in any known natural protein. Native proteins containing alpha-strand regions or alpha-sheet-patterned hydrogen bonding include synaptotagmin, lysozyme, and potassium channels, where the alpha-strands line the ion-conducting pore.[4]
Evidence for the existence of alpha-sheet in a mutant form of transthyretin has been presented.[7] Alpha-sheet conformations have been observed in crystal structures of short non-natural peptides, especially those containing a mixture of L and D amino acids. The first crystal structure containing an alpha sheet was observed in the capped tripeptide Boc–AlaL–a-IleD–IleL–OMe.[8] Other peptides that assume alpha-sheet structures include capped diphenyl-glycine-based dipeptides[9] and tripeptides.[10]
Role in amyloidogenesis
The alpha sheet has been proposed as a possible intermediate state in the conformational change in the formation of amyloid fibrils by peptides and proteins such as amyloid beta, poly-glutamine repeats, lysozyme, prion proteins, and transthyretin repeats, all of which are associated with protein misfolding disease. For example, amyloid beta is a major component of amyloid plaques in the brains of Alzheimer's disease patients,[6] and polyglutamine repeats in the huntingtin protein are associated with Huntington's disease.[11] These proteins undergo a conformational change from largely random coil or alpha helix structures to the highly ordered beta sheet structures found in amyloid fibrils. Most beta sheets in known proteins are "twisted" about 15° for optimal hydrogen bonding and steric packing; however, some evidence from electron crystallography suggests that at least some amyloid fibrils contain "flat" sheets with only 1–2.5° of twist.[12] An alpha-sheet amyloid intermediate is suggested to explain some anomalous features of the amyloid fibrillization process, such as the evident amino acid sequence dependence of amyloidogenesis despite the belief that the amyloid fold is mainly stabilized by the protein backbone.[13][14]
Xu,[15] using atomic force microscopy, has shown that formation of amyloid fibers is a two-step process in which proteins first aggregate into colloidal spheres of ≈20 nm diameter. The spheres then join together spontaneously to form linear chains, which evolve into mature amyloid fibers. The formation of these linear chains appears to be driven by the development of an electrostatic dipole in each of the colloidal spheres strong enough to overcome coulomb repulsion. This suggests a possible mechanism by which alpha sheet may promote amyloid aggregation; the peptide bond has a relatively large intrinsic electrostatic dipole, but normally the dipoles of nearby bonds cancel each other out. In the alpha sheet, unlike other conformations, the peptide bonds are oriented in parallel so that the dipoles of the individual bonds can add up to create a strong overall electrostatic dipole.
Notably, the protein lysozyme is among the few native-state proteins shown to contain an alpha-strand region; lysozyme from both chickens and humans contains an alpha strand located close to the site of a mutation known to cause hereditary amyloidosis in humans, usually an autosomal dominant genetic disease.[4] Molecular dynamics simulations of the mutant protein reveal that the region around the mutation assumes an alpha strand conformation.[6] Lysozyme is among the naturally occurring proteins known to form amyloid fibers under experimental conditions, and both natively alpha-strand region and the mutation site fall within the larger region identified as the core of lysozyme amyloid fibrillogenesis.[16][17]
A mechanism for direct alpha sheet and beta sheet interconversion has also been suggested, based on peptide plane flipping in which the αRαL dipeptide inverts to produce a ββ dihedral angle conformation. This process has also been observed in simulations of transthyretin[18] and implicated as occurring naturally in certain protein families by examination of their dihedral angle conformations in crystal structures.[19][20] It is suggested that alpha-sheet folds into multi-strand solenoids.[21]
Evidence employing retro-enantio N-methylated peptides, or those with alternating L and D amino acids, as inhibitors of beta-amyloid aggregation is consistent with alpha-sheet being the main material of the amyloid precursor.[22][23][24] [25][26]
References
- Pauling, L. & Corey, R. B. (1951). The pleated sheet, a new layer configuration of polypeptide chains. Proc. Natl. Acad. Sci. USA 37, 251–6. PMID 14834147
- Pauling, L. & Corey, R. B. (1951). The structure of feather rachis keratin. Proc. Natl. Acad. Sci. USA 37, 256–261. doi:10.1073/pnas.37.5.256 PMID 14834148
- Pauling, L. & Corey, R. B. (1951). Configurations of Polypeptide Chains With Favored Orientations Around Single Bonds: Two New Pleated Sheets. Proc. Natl. Acad. Sci. USA 37, 729–740. PMID 16578412
- Daggett V. (2006). Alpha-sheet: The toxic conformer in amyloid diseases? Acc Chem Res 39(9):594-602. doi:10.1021/ar0500719 PMID 16981675
- Babin V, Roland C, Sagui C. (2011). The alpha-sheet: A missing in action secondary structure. Proteins 79:937-946. doi:10.1002/prot.22935
- Armen RS, DeMarco ML, Alonso DO, Daggett V. (2011). Pauling and Corey's alpha-pleated sheet structure may define the prefibrillar amyloidogenic intermediate in amyloid disease. Proc Natl Acad Sci USA 101(32):11622-7. doi:10.1073/pnas.0401781101 PMID 15280548
- Hilaire MR, Ding B, Mukherjee D, Chen J, Gai F. (2018). Possible existence of alpha-sheets in the amyloid fibrils formed by a mutant form of a peptide from transthyretin. Journal of the American Chemical Society 140:629-635. doi:10.1021/jacs.7b09262
- Di Blasio B, Saviano M, Fattorusso R, Lombardi A, Pedone C, Valle V, Lorenzi GP. (1994). A crystal structure with features of an antiparallel alpha-pleated sheet. Biopolymers 34(11):1463-8. PMID 7827259
- De Simone G, Lombardi A, Galdiero S, Nastri F, Di Costanzo L, Gohda S, Sano A, Yamada T, Pavone V. (2000). The crystal structure of a Dcp-containing peptide. Biopolymers 53(2):182-8. PMID 10679622
- Pavone V, Lombardi A, Saviano M, Nastri F, Zaccaro L, Maglio O, Pedone C, Omote Y, Yamanaka Y, Yamada T. (1998). Conformational behaviour of C(alpha,alpha)-diphenylglycine: folded vs. extended structures in DphiG-containing tripeptides. 4(1):21-32. PMID 9523753
- Armen RS, Bernard BM, Day R, Alonso DO, Daggett V. (2005). Characterization of a possible amyloidogenic precursor in glutamine-repeat neurodegenerative diseases. Proc Natl Acad Sci USA 102(38):13433-8. PMID 16157882
- Jimenez, J. L., Nettleton, E. J., Bouchard, M., Robinson, C. V., Dobson, C. M. & Saibil, H. R. (2002). The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. USA 99, 9196–9201. PMID 12093917
- Fraser, P. E., Duffy, L. K., O'Mally, M. B., Nguyen, J., Inouye, H. & Kirschner, D. A. (1991). Morphology and antibody recognition of synthetic beta-amyloid peptides. J. Neurosci. Res. 28, 474–485. PMID 1908024.
- Malinchik, S. B., Inouye, H., Szumowski, K. E. & Kirschner, D. A. (1998). Structural analysis of Alzheimer's beta(1-40) amyloid: protofilament assembly of tubular fibrils. Biophys. J. 74, 537–545. PMID 9449354
- Xu S. Aggregation drives "misfolding" in protein amyloid fiber formation. Amyloid 2007 Jun;14(2):119-31. PMID 17577685
- Frare, D.; Polverino de Laureto, P.; Zurdo, J.; Dobson, C. M., Fontana, A. (2004). A highly amyloidogenic region of hen lysozyme. J Mol Biol 340: 1153-1165. PMID 15236974
- Frare E, Mossuto MF, Polverino de Laureto P, Dumoulin M, Dobson CM, Fontana A. (2006). Identification of the core structure of lysozyme amyloid fibrils by proteolysis. J Mol Biol 361(3):551-61. PMID 16859705
- Yang MF, Lei M, Yordanov B, Huo SH. (2006). Peptide plane can flip in two opposite directions: implication in amyloid formation of transthyretin. J Phys Chem B 110(12):5829-33. PMID 16553385
- Milner-White EJ, Watson JD, Qi G, Hayward S. (2006). Amyloid formation may involve alpha- to beta sheet interconversion via peptide plane flipping. Structure 14(9):1369-76. PMID 16962968
- Hayward S, Milner-White EJ. (2008). The geometry of alpha-sheet: Implications for its possible function as amyloid precursor in proteins Proteins 71:425-431. PMID 17957773
- Hayward S, Milner-White EJ. (2011). Simulation of the β- to α-sheet transition results in a twisted sheet for antiparallel and an α-nanotube for parallel strands: implications for amyloid format. Proteins 79(11) 3193-3207. PMID 21989939
- Grillo-Bosch D, Carulla N, Cruz M, Pujol-Pina R, Madurga S, Rabanal F, Giralt E. (2009). Retro-enantio N-methylated peptides as inhibitors of beta-amyloid aggregation. ChemMedChem 4, 1488-1494. PMID 19591190
- Kellock J, Hopping G, Cauchey B, Daggett V. (2016). Peptides composed of alternating L- and D-amino acids inhibit amyloidogenesis in three distinct amyloid systems independent of sequence. Journal of Molecular Biology 428, 2317-2328
- Paranjapye N, Daggett V. (2018). De novo designed alpha-sheet peptides inhibit functional amyloid formation of Streptococcus mutant biofilms. Journal of Molecular Biology 430, 3764-3773
- Maris NL, Shea D. (2018). Chemical and Physical Variability in Structural Isomers of an L/D α‑Sheet Peptide Designed To Inhibit Amyloidogenesis. Biochemistry 57, 507-510. doi:10.1021/acs.biochem.7b00345
- Shea D, Hsu C-C. (2019). α-Sheet secondary structure in amyloid β-peptide drives aggregation and toxicity in Alzheimer’s disease. Proc Nat Acad Sci USA 116(18) 8895-8900. doi:10.1073/pnas.1820585116