Activity-based proteomics
Activity-based proteomics, or activity-based protein profiling (ABPP) is a functional proteomic technology that uses chemical probes that react with mechanistically related classes of enzymes.[1]
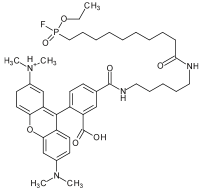
Description
The basic unit of ABPP is the probe, which typically consists of two elements: a reactive group (RG, sometimes called a "warhead") and a tag. Additionally, some probes may contain a binding group which enhances selectivity. The reactive group usually contains a specially designed electrophile that becomes covalently-linked to a nucleophilic residue in the active site of an active enzyme. An enzyme that is inhibited or post-translationally modified will not react with an activity-based probe. The tag may be either a reporter such as a fluorophore or an affinity label such as biotin or an alkyne or azide for use with the Huisgen 1,3-dipolar cycloaddition (also known as click chemistry).[2]
Advantages
A major advantage of ABPP is the ability to monitor the availability of the enzyme active site directly, rather than being limited to protein or mRNA abundance. With classes of enzymes such as the serine hydrolases [3] and metalloproteases [4] that often interact with endogenous inhibitors or that exist as inactive zymogens, this technique offers a valuable advantage over traditional techniques that rely on abundance rather than activity.
Multidimensional protein identification technology
In recent years ABPP has been combined with tandem mass spectrometry enabling the identification of hundreds of active enzymes from a single sample. This technique, known as ABPP-MudPIT (multidimensional protein identification technology) is especially useful for profiling inhibitor selectivity as the potency of an inhibitor can be tested against hundreds of targets simultaneously.
ABPP were first reported in the 1990s in the study of proteases.[5][6]
See also
- Mass spectrometry
- Proteomics
- Related inhibitors MAFP and DIFP
References
- Berger AB, et al. Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. American Journal of Pharmacogenomics 2004 Article
- Speers AE, et al. Activity-Based Protein Profiling in Vivo Using a Copper(I)-Cataqlyzed Azide-Alkyne [3+2] Cycloaddition Journal of the American Chemical Society 2003 Article
- Liu Y, et al. Activity-based protein profiling: The serine hydrolases Proceedings of the National Academy of Sciences 1999 Article
- Saghatelian A , et al. Activity-based probes for the proteomic profiling of metalloproteases Proceedings of the National Academy of Sciences 2004 Article
- Kam CM, et al. Biotinylated isocoumarins, new inhibitors and reagents for detection, localization, and isolation of serine proteases. Bioconjugate chemistry 1993
- Abuelyaman AS, et al. Fluorescent derivatives of diphenyl [1-(N-peptidylamino)alkyl]phosphonate esters: synthesis and use in the inhibition and cellular localization of serine proteases. Bioconjugate chemistry 1994