SUHW4
Zinc finger protein 280D, also known as Suppressor Of Hairy Wing Homolog 4, SUWH4, Zinc Finger Protein 634, ZNF634, or KIAA1584, is a protein that in humans is encoded by the ZNF280D gene located on chromosome 15q21.3.[4][5]
| ZNF280D | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | ZNF280D, SUHW4, ZNF634, zinc finger protein 280D | ||||||||||||||||||||||||
| External IDs | MGI: 2384583 HomoloGene: 17151 GeneCards: ZNF280D | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl |
| ||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | n/a | Chr 9: 72.27 – 72.36 Mb | |||||||||||||||||||||||
| PubMed search | [2] | [3] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Gene
There are a total of 24 possible exons in any variant of the ZNF280D gene.[6] ZNF280D is oriented on the minus strand of Chromosome 15 and spans 288.396 kb.[7] Surrounding genes at the same locus include TEX9, HMGB1P33, MNS1, LOC645877, and LOC145783.[8]
mRNA
At least 24 spliced variants have been identified.[9] There are 7 probable alternative promoters. The mRNAs appear to differ by truncation of the 5' end, truncation of the 3' end, presence or absence of 12 cassette exons, overlapping exons with different boundaries, splicing versus retention of 5 introns.[9] The longest splice form contains 4428 bp.[10]
Protein
Composition and Domains
The ZNF280D protein is 979 amino acids in length.[11] The protein contains a domain of unknown function (DUF4195) spanning from amino acid 45 to amino acid 230.[11] DUF4195 (pfam13826) is a family that is found at the N-terminus of metazoan proteins that carry PHD-like zinc-finger domains; the function is unknown.[12] ZNF280D protein also contains five highly conserved Cys2His2-type zinc finger domains.[13] Zinc fingers have the ability to bind DNA, which supports the speculative role of ZNF280D as a transcription factor.[14] The protein has a weight of approximately 109.3 kdal.[15] Charge cluster analysis reveals a negative charge cluster near the N-terminus from amino acids 16-43.[15] Charge clusters are associated with functional domains of cellular transcription factors, providing further support for ZNF280D as a possible transcription factor.[16]
Interactions
ZNF280D has been experimentally determined to interact with CBX5 and CBX3 proteins.[17] These proteins both play a role in the formation of heterochromatin, which presents a possible functional role of ZNF280D as a transcriptional repressor.[18][19]
SNPs
There are a number of SNPs that have been observed in the human population.[20] The image below lists some of the most frequently occurring.
Regulation
mRNA Level

A number of transcription factors are predicted to bind to the predicted promoter region.[21]
Protein Level
ZNF280D protein contains 66 serine, 17 threonine, and 6 tyrosine residues all of which are potential phosphorylation sites.[22]
The glycine residue at position 2 is a probable candidate for N-terminal acetylation.[23] There are seven probable sumoylation sites.[24]
Expression
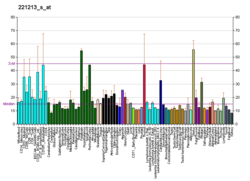
ZNF280D is ubiquitously expressed at relatively low levels throughout almost all tissues in the human body.[25]
In one study, the expression of ZNF280D was compared between endothelial progenitor cells in cord blood and peripheral blood. The results show that expression was significantly higher in cord blood. This supports a possible involvement of ZNF280D in embryonic development or cell differentiation.[26]
Evolution
A number of orthologs and distant homologs have been identified for the human ZNF280D protein. There are also four paralogs to ZNF280D in the human genome.[27]
| Protein Name | Species | Date of Divergence (Million Years Ago)[28] | Accession Number | Sequence Length (# amino acids) | Sequence Identity | E Value |
|---|---|---|---|---|---|---|
| ZNF280D | Mouse (Mus musculus) | 92.3 | NP_666336 | 974 | 76% | 0 |
| ZNF280D Isoform X1 | Cow (Bos taurus) | 94.2 | XP_002690904 | 974 | 88% | 0 |
| ZNF280D | African bush elephant (Loxodonta africana) | 98.7 | XP_003418437 | 979 | 90% | 0 |
| ZNF280D Isoform X1 | Chinese softshell turtle (Pelodiscus sinensis) | 296 | XP_006112544 | 993 | 61% | 0 |
| ZNF280D | Saker falcon (Falco cherrug) | 296 | XP_005435548 | 939 | 60% | 0 |
| ZNF280D | Ground tit (Pseudopdoces humilis) | 296 | XP_005521527 | 936 | 57% | 0 |
| ZNF280D Isoform X1 | Chinese alligator (Alligator sinensis) | 296 | XP_006018037 | 277 | 50% | 1E-64 |
| ZNF280D | Western clawed frog (Xenopus tropicalis) | 371.2 | XP_002940298 | 1071 | 49% | 0 |
| ZNF280D Isoform X1 | Zebrafish (Danio rerio) | 400.1 | XP_005166581 | 879 | 43% | 2E-172 |
| ZNF280D-like | Acorn worm (Saccoglossus kowalevskii) | 661.2 | XP_006811912 | 733 | 32% | 5E-50 |
| Zinc Finger Protein 36 | Sea squirt (Ciona intestinalis) | 722.5 | NP_001041459 | 581 | 32% | 7E-58 |
| GL11474 | Fruit fly (Drosophila persimilis) | 782.7 | XP_002016218 | 1271 | 29% | 5E-16 |
| Zinc Finger Protein | Eye worm (Loa loa) | 937.5 | XP_003139663 | 495 | 33% | 6E-19 |
| Zap1p | Yeast (Saccharomyces cerevisiae S288c) | 12158 | NP_012479 | 880 | 33% | 2E-7 |
| Paralog Name | Accession Number | Sequence Length (# amino acids) | Sequence Identity | E Value |
|---|---|---|---|---|
| ZNF280C | NP_060136 | 737 | 68% | 0 |
| ZNF280B | NP_542942 | 543 | 54% | 0 |
| ZNF280A | NP_542778 | 542 | 49% | 6E-148 |
| ZNF280E (pogo transposable element with ZNF domain isoform 1) | NP_055915 | 1410 | 44% | 4E-134 |
References
- GRCm38: Ensembl release 89: ENSMUSG00000038535 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Nagase T, Kikuno R, Nakayama M, Hirosawa M, Ohara O (August 2000). "Prediction of the coding sequences of unidentified human genes. XVIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Research. 7 (4): 273–81. doi:10.1093/dnares/7.4.271. PMID 10997877.
- "Entrez Gene: SUHW4 suppressor of hairy wing homolog 4 (Drosophila)".
- "Genetic Testing Registry". NCBI. Retrieved 7 May 2014.
- "GeneCards". Retrieved 7 May 2014.
- "ZNF280D zinc finger protein 280D [ Homo sapiens (human) ]". NCBI. Retrieved 9 May 2014.
- "Homo sapiens complex locus ZNF280D, encoding zinc finger protein 280D and hypothetical LOC145783". NCBI AceView. Retrieved 7 May 2014.
- "Homo sapiens zinc finger protein 280D, mRNA (cDNA clone MGC:168023 IMAGE:9020400), complete cds". NCBI. Retrieved 7 May 2014.
- "Zinc finger protein 280D [Homo sapiens]". NCBI. Retrieved 7 May 2014.
- "pfam13836: DUF4195". NCBI. Retrieved 7 May 2014.
- "Q6N043 (Z280D_HUMAN)". UniProt. Retrieved 7 May 2014.
- Klug A (October 1999). "Zinc finger peptides for the regulation of gene expression". Journal of Molecular Biology. 293 (2): 215–8. doi:10.1006/jmbi.1999.3007. PMID 10529348.
- Brendel, Voker. "SAPS Statistical Analysis of PS". SDSC Workbench. Retrieved 7 May 2014.
- Brendel V, Karlin S (August 1989). "Association of charge clusters with functional domains of cellular transcription factors". Proceedings of the National Academy of Sciences of the United States of America. 86 (15): 5698–702. doi:10.1073/pnas.86.15.5698. PMC 297697. PMID 2569737.
- "STRING input ZNF280D (Homo sapiens)". STRING. Retrieved 10 May 2014.
- "CBX5 [Homo sapiens]". NCBI. Retrieved 10 May 2014.
- "CBX3 [Homo sapiens]". NCBI. Retrieved 10 May 2014.
- "SNP linked to Gene ZNF280D(geneID:54816) Via Contig Annotation". NCBI. Retrieved 10 May 2014.
- "Genomatix". Genomatix. Retrieved 7 May 2014.
- Blom N, Gammeltoft S, Brunak S (December 1999). "Sequence and structure-based prediction of eukaryotic protein phosphorylation sites". Journal of Molecular Biology. 294 (5): 1351–62. doi:10.1006/jmbi.1999.3310. PMID 10600390.
- Kiemer, Lars. "NetAcet: Prediction of N-terminal acetylation sites". NetAcet. Retrieved 7 May 2014.
- "SUMOplot™ Analysis Program". Retrieved 7 May 2014.
- "GDS596 / 221213_s_at / ZNF280D". GEO Profile. NCBI. Retrieved 7 May 2014.
- "ZNF280D – Endothelial progenitor cells from cord blood and adult peripheral blood". NCBI. Retrieved 9 May 2014.
- "NCBI Basic Local Alignment Search Tool". NCBI. Retrieved 10 May 2014.
- "TimeTree". TimeTree. Retrieved 10 May 2014.
Further reading
- Mehrle A, Rosenfelder H, Schupp I, del Val C, Arlt D, Hahne F, Bechtel S, Simpson J, Hofmann O, Hide W, Glatting KH, Huber W, Pepperkok R, Poustka A, Wiemann S (January 2006). "The LIFEdb database in 2006". Nucleic Acids Research. 34 (Database issue): D415-8. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.
- Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, Bechtel S, Sauermann M, Korf U, Pepperkok R, Sültmann H, Poustka A (October 2004). "From ORFeome to biology: a functional genomics pipeline". Genome Research. 14 (10B): 2136–44. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Böcher M, Blöcker H, Bauersachs S, Blum H, Lauber J, Düsterhöft A, Beyer A, Köhrer K, Strack N, Mewes HW, Ottenwälder B, Obermaier B, Tampe J, Heubner D, Wambutt R, Korn B, Klein M, Poustka A (March 2001). "Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs". Genome Research. 11 (3): 422–35. doi:10.1101/gr.GR1547R. PMC 311072. PMID 11230166.
- Hartley JL, Temple GF, Brasch MA (November 2000). "DNA cloning using in vitro site-specific recombination". Genome Research. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Bonaldo MF, Lennon G, Soares MB (September 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.