U6 spliceosomal RNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (small nuclear ribonucleoprotein), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes.
| U6 spliceosomal RNA | |
|---|---|
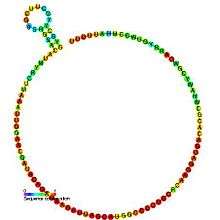 Predicted secondary structure and sequence conservation of U6 | |
| Identifiers | |
| Symbol | U6 |
| Rfam | RF00026 |
| Other data | |
| RNA type | Gene; snRNA; splicing |
| Domain(s) | Eukaryota |
| GO | 0000351 0000353 0030621 0005688 0046540 |
| SO | 0000396 |
| PDB structures | PDBe |
The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome,[1] suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution.
It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes.[2] This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability.
The U6 snRNA gene has been isolated in many organisms,[3] including C. elegans.[4] Among them, baker's yeast (Saccharomyces cerevisiae) is a commonly used model organism in the study of snRNAs.
The structure and catalytic mechanism of U6 snRNA resembles that of domain V of group II introns.[5][6] The formation of the triple helix in U6 snRNA is deemed to be important in splicing activity, where its role is to bring the catalytic site to the splice site.[6]
Role
Base-pair specificity of the U6 snRNA allows the U6 snRNP to bind tightly to the U4 snRNA and loosely to the U5 snRNA of a triple-snRNP during the initial phase of the splicing reaction. As the reaction progresses, the U6 snRNA is unzipped from U4 and binds to the U2 snRNA. At each stage of this reaction, the U6 snRNA secondary structure undergoes extensive conformational changes.[7]
The association of U6 snRNA with the 5' end of the intron via base-pairing during the splicing reaction occurs prior to the formation of the lariat (or lasso-shaped) intermediate, and is required for the splicing process to proceed. The association of U6 snRNP with U2 snRNP via base-pairing forms the U6-U2 complex, a structure that comprises the active site of the spliceosome.[8]:433–437
Secondary structure
While the putative secondary structure consensus base pairing is confined to a short 5' stem-loop, much more extensive structures have been proposed for specific organisms such as in yeast.[9] In addition to the 5' stem loop, all confirmed U6 snRNAs can form the proposed 3' intramolecular stem loop.[10]
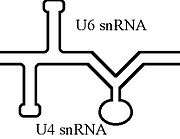
The U6 snRNA is known to form an extensive base-pair interactions with U4 snRNA.[11] This interaction has been shown to be mutually exclusive to that of the 3' intramolecular stem loop.[7]
Associated proteins
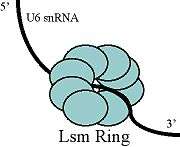
Free U6 snRNA is found to be associated with the proteins Prp24and the LSms. Prp24 is thought to form an intermediate complex with the U6 snRNA, in order to facilitate the extensive base-pairing between the U4 and U6 snRNAs, and the Lsms may aid in Prp24 binding. The approximate location of these protein binding domains was determined, and the proteins were later visualized by electron microscopy. This study suggests that in the free form of U6, Prp24 binds to the telestem and the uradine-rich 3' tail of the U6 snRNA is threaded through the ring of Lsms. Another important NTC-related protein associated with U6 is Cwc2, which by interaction with important catalytic RNA elements induces the formation of a functional catalytic core in the spliceosome. Cwc2 and U6 achieve formation of this complex by interaction with the ISL and regions located near the 5' splice site.[12]
See also
References
- Brow DA, Guthrie C (July 1988). "Spliceosomal RNA U6 is remarkably conserved from yeast to mammals". Nature. 334 (6179): 213–8. Bibcode:1988Natur.334..213B. doi:10.1038/334213a0. PMID 3041282. S2CID 4236176.
- Marz M, Kirsten T, Stadler PF (December 2008). "Evolution of spliceosomal snRNA genes in metazoan animals". Journal of Molecular Evolution (Submitted manuscript). 67 (6): 594–607. Bibcode:2008JMolE..67..594M. doi:10.1007/s00239-008-9149-6. PMID 19030770. S2CID 18830327.
- Anderson MA, Purcell J, Verkuijl SA, Norman VC, Leftwich PT, Harvey-Samuel T, Alphey LS (March 2020). "In Vitro Validation of Pol III Promoters". ACS Synthetic Biology. 9 (3): 678–681. doi:10.1021/acssynbio.9b00436. PMC 7093051. PMID 32129976.
- Thomas J, Lea K, Zucker-Aprison E, Blumenthal T (May 1990). "The spliceosomal snRNAs of Caenorhabditis elegans". Nucleic Acids Research. 18 (9): 2633–42. doi:10.1093/nar/18.9.2633. PMC 330746. PMID 2339054.
- Toor N, Keating KS, Taylor SD, Pyle AM (April 2008). "Crystal structure of a self-spliced group II intron". Science. 320 (5872): 77–82. Bibcode:2008Sci...320...77T. doi:10.1126/science.1153803. PMC 4406475. PMID 18388288.
- Fica SM, Mefford MA, Piccirilli JA, Staley JP (May 2014). "Evidence for a group II intron-like catalytic triplex in the spliceosome". Nature Structural & Molecular Biology. 21 (5): 464–471. doi:10.1038/nsmb.2815. PMC 4257784. PMID 24747940.
- Fortner DM, Troy RG, Brow DA (January 1994). "A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing". Genes & Development. 8 (2): 221–33. doi:10.1101/gad.8.2.221. PMID 8299941.
- Weaver, Robert J. (2008). Molecular Biology. Boston: McGraw Hill Higher Education. ISBN 978-0-07-127548-4.
- Karaduman R, Fabrizio P, Hartmuth K, Urlaub H, Lührmann R (March 2006). "RNA structure and RNA-protein interactions in purified yeast U6 snRNPs". Journal of Molecular Biology. 356 (5): 1248–62. doi:10.1016/j.jmb.2005.12.013. hdl:11858/00-001M-0000-0012-E5F7-6. PMID 16410014.
- Butcher SE, Brow DA (June 2005). "Towards understanding the catalytic core structure of the spliceosome". Biochemical Society Transactions. 33 (Pt 3): 447–9. doi:10.1042/BST0330447. PMID 15916538.
- Orum H, Nielsen H, Engberg J (November 1991). "Spliceosomal small nuclear RNAs of Tetrahymena thermophila and some possible snRNA-snRNA base-pairing interactions". Journal of Molecular Biology. 222 (2): 219–32. doi:10.1016/0022-2836(91)90208-N. PMID 1960724.
- Rasche N, Dybkov O, Schmitzová J, Akyildiz B, Fabrizio P, Lührmann R (March 2012). "Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre". The EMBO Journal. 31 (6): 1591–604. doi:10.1038/emboj.2011.502. PMC 3321175. PMID 22246180.
Further reading
- Zwieb C (January 1997). "The uRNA database". Nucleic Acids Research. 25 (1): 102–3. doi:10.1093/nar/25.1.102. PMC 146409. PMID 9016512.