Small conditional RNA
A small conditional RNA (scRNA) is a small RNA molecule or complex (typically less than approximately 100 nt) engineered to interact and change conformation conditionally in response to cognate molecular inputs so as to perform signal transduction in vitro, in situ, or in vivo.[1] In the absence of cognate input molecules, scRNAs are engineered to coexist metastably or stably without interacting. Detection of the cognate inputs initiates downstream conformational changes of one or more scRNAs leading to generation of the desired output signal. The output signal may be intended to read out the state of endogenous biological circuitry (e.g., mapping gene expression for biological research or medical diagnosis),[2] or to regulate the state of endogenous biological circuitry (e.g., perturbing gene expression for biological research or medical treatment).[1] scRNA sequences can be programmed to recognize different inputs or to activate different outputs,[1][2][3] achieving even single-nucleotide sequence selectivity.[3] scRNA signal transduction exploits principles from the emerging disciplines of dynamic RNA nanotechnology, molecular programming, and synthetic biology.
_mechanism.png)
Examples of scRNA signal transduction
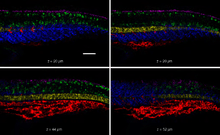
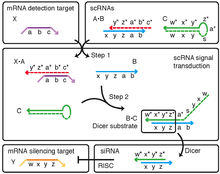
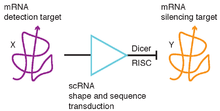
Fluorophore-labeled scRNAs have been engineered to transduce between detection of mRNA targets and generation of bright fluorescent amplification polymers in situ (Figure 1).[2] In this context, scRNA signal transduction enables multiplexed mapping of mRNA expression within intact vertebrate embryos (Figure 2).[2] scRNAs have been engineered to perform shape and sequence transduction to conditionally produce a Dicer substrate targeting 'silencing target' mRNA Y upon detection of an independent 'detection target' mRNA X, with subsequent Dicer processing yielding a small interfering RNA (siRNA) targeting mRNA Y for destruction (Figure 3).[1] In this context, scRNA signal transduction provides a step towards implementing conditional RNA interference (Figure 4).
Design elements
scRNAs can be engineered to exploit diverse design elements:[1]
- scRNA reactants may be metastable or stable
- scRNA signal transduction cascades can be catalytic or non-catalytic
- scRNAs can nucleate with each other or with input or output molecules via toehold/toehold nucleation, loop/toehold nucleation, or template/toehold nucleation
- scRNAs can dissociate with each other or with input or output molecules via 3-way branch migration, 4-way branch migration, or spontaneous dissociation
- scRNA reactants can be monomers, dimers, or other multimers
References
- Hochrein, L.M.; Schwarzkopf, M.; Shahgholi, M.; Yin, P.; Pierce, N.A. (2013). "Conditional Dicer substrate formation via shape and sequence transduction with small conditional RNAs". Journal of the American Chemical Society. 135 (46): 17322–17330. doi:10.1021/ja404676x. PMC 3842090. PMID 24219616.
- H.M.T. Choi, J.Y. Chang, L.A. Trinh, J.E. Padilla, S.E. Fraser, and N.A. Pierce. Choi, H.M.T.; Chang, J.Y.; Trinh, L.A.; Padilla, J.E.; Fraser, S.E.; Pierce, N.A. (2010). "Programmable in situ amplification for multiplexed imaging of mRNA expression". Nature Biotechnology. 28 (11): 1208–1212. doi:10.1038/nbt.1692. PMC 3058322. PMID 21037591.
- Sternberg, J.B.; Pierce., N.A. (2014). "Exquisite sequence selectivity with small conditional RNAs". Nano Letters. 14 (8): 4568–4572. doi:10.1021/nl501593r. PMC 4134187. PMID 24979041.