Secondary plot (kinetics)
In enzyme kinetics, a secondary plot uses the intercept or slope from several Lineweaver–Burk plots to find additional kinetic constants.[1][2]
For example, when a set of v by [S] curves from an enzyme with a ping–pong mechanism (varying substrate A, fixed substrate B) are plotted in a Lineweaver–Burk plot, a set of parallel lines will be produced.
The following Michaelis–Menten equation relates the initial reaction rate v0 to the substrate concentrations [A] and [B]:
The y-intercept of this equation is equal to the following:
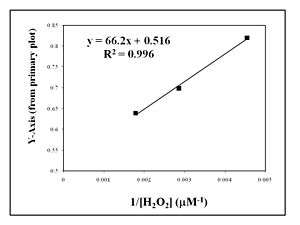
The y-intercept is determined at several different fixed concentrations of substrate B (and varying substrate A). The y-intercept values are then plotted versus 1/[B] to determine the Michaelis constant for substrate B, , as shown in the Figure to the right.[3] The slope is equal to divided by and the intercept is equal to 1 over .
Secondary plot in inhibition studies
A secondary plot may also be used to find a specific inhbition constant, KI.
For a competitive enzyme inhibitor, the apparent Michaelis constant is equal to the following:
The slope of the Lineweaver-Burk plot is therefore equal to:
If one creates a secondary plot consisting of the slope values from several Lineweaver-Burk plots of varying inhibitor concentration [I], the competitive inhbition constant may be found. The slope of the secondary plot divided by the intercept is equal to 1/KI. This method allows one to find the KI constant, even when the Michaelis constant and vmax values are not known.
References
- A. Cornish-Bowden. Fundamentals of Enzyme kinetics Rev. ed., Portland: London, England, (1995) pp. 30-37, 56-57.
- J. N. Rodriguez-Lopez, M. A. Gilabert, J. Tudela, R. N. F. Thorneley, and F. Garcia-Canovas. Biochemistry, 2000, 39, 13201-13209.
- The Horseradish Peroxidase/ o-Phenylenediamine (HRP/OPD) System Exhibits a Two-Step Mechanism. M. K. Tiama and T. M. Hamilton, Journal of Undergraduate Chemistry Research, 4, 1 (2005).