SEP15
15 kDa selenoprotein is a protein that in humans is encoded by the SEP15 gene.[5] Two alternatively spliced transcript variants encoding distinct isoforms have been found for this gene.
| SELENOF | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | SELENOF, selenoprotein F, SEP15 | ||||||||||||||||||||||||
| External IDs | OMIM: 606254 MGI: 1927947 HomoloGene: 3145 GeneCards: SELENOF | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 1: 86.86 – 86.91 Mb | Chr 3: 144.57 – 144.6 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Function
This gene encodes a selenoprotein, which contains a selenocysteine (Sec) residue at its active site. The selenocysteine is encoded by the UGA codon that normally signals translation termination. The 3' UTR of selenoprotein genes have a common stem-loop structure, the sec insertion sequence (SECIS), that is necessary for the recognition of UGA as a Sec codon rather than as a stop signal. Studies in mouse suggest that this selenoprotein may have redox function and may be involved in the quality control of protein folding.[5]
Clinical significance
This gene is localized on chromosome 1p31, a genetic locus commonly mutated or deleted in human cancers.[5]
Protein domain
| Sep15 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
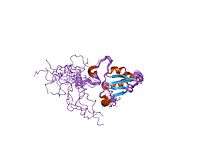 Solution structure of SelM from Mus musculus | |||||||||
| Identifiers | |||||||||
| Symbol | Sep15_SelM | ||||||||
| Pfam | PF08806 | ||||||||
| InterPro | IPR014912 | ||||||||
| |||||||||
The protein this gene encodes for is often called Sep15 however in the case of mice, it is named SelM. This protein is a selenoprotein only found in eukaryotes. This domain has a thioredoxin-like domain and a surface accessible active site redox motif.[6] This suggests that they function as thiol-disulfide isomerases involved in disulfide bond formation in the endoplasmic reticulum.[6]
Function
Recent studies have shown in mice, where the SEP15 gene has been silenced the mice subsequently became deficient in SEP15 and were able to inhibit the development of colorectal cancer.[7]
Structure
The particular structure has an alpha/beta central domain which is actually made up of three alpha helices and a mixed parallel/anti-parallel four-stranded beta-sheet.[6]
References
- GRCh38: Ensembl release 89: ENSG00000183291 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000037072 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: SEP15 15 kDa selenoprotein".
- Ferguson AD, Labunskyy VM, Fomenko DE, Araç D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J (February 2006). "NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family". The Journal of Biological Chemistry. 281 (6): 3536–43. doi:10.1074/jbc.M511386200. PMID 16319061.
- Tsuji PA, Naranjo-Suarez S, Carlson BA, Tobe R, Yoo MH, Davis CD (September 2011). "Deficiency in the 15 kDa selenoprotein inhibits human colon cancer cell growth". Nutrients. 3 (9): 805–17. doi:10.3390/nu3090805. PMC 3257736. PMID 22254125.
Further reading
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Gladyshev VN, Jeang KT, Wootton JC, Hatfield DL (April 1998). "A new human selenium-containing protein. Purification, characterization, and cDNA sequence". The Journal of Biological Chemistry. 273 (15): 8910–5. doi:10.1074/jbc.273.15.8910. PMID 9535873.
- Kumaraswamy E, Malykh A, Korotkov KV, Kozyavkin S, Hu Y, Kwon SY, Moustafa ME, Carlson BA, Berry MJ, Lee BJ, Hatfield DL, Diamond AM, Gladyshev VN (November 2000). "Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology". The Journal of Biological Chemistry. 275 (45): 35540–7. doi:10.1074/jbc.M004014200. PMID 10945981.
- Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Böcher M, Blöcker H, Bauersachs S, Blum H, Lauber J, Düsterhöft A, Beyer A, Köhrer K, Strack N, Mewes HW, Ottenwälder B, Obermaier B, Tampe J, Heubner D, Wambutt R, Korn B, Klein M, Poustka A (March 2001). "Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs". Genome Research. 11 (3): 422–35. doi:10.1101/gr.GR1547R. PMC 311072. PMID 11230166.
- Korotkov KV, Kumaraswamy E, Zhou Y, Hatfield DL, Gladyshev VN (May 2001). "Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells". The Journal of Biological Chemistry. 276 (18): 15330–6. doi:10.1074/jbc.M009861200. PMID 11278576.
- Kumaraswamy E, Korotkov KV, Diamond AM, Gladyshev VN, Hatfield DL (2002). "Genetic and functional analysis of mammalian Sep15 selenoprotein". Protein Sensors and Reactive Oxygen Species - Part A: Selenoproteins and Thioredoxin. Methods in Enzymology. 347. pp. 187–97. doi:10.1016/S0076-6879(02)47017-6. ISBN 978-0-12-182248-4. PMID 11898406.
- Wu HJ, Lin C, Zha YY, Yang JG, Zhang MC, Zhang XY, Liang X, Fu M, Wu M (February 2003). "[Redox reactions of Sep15 and its relationship with tumor development]". AI Zheng = Aizheng = Chinese Journal of Cancer. 22 (2): 119–22. PMID 12600282.
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J (May 2003). "Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides". Nature Biotechnology. 21 (5): 566–9. doi:10.1038/nbt810. PMID 12665801.
- Apostolou S, Klein JO, Mitsuuchi Y, Shetler JN, Poulikakos PI, Jhanwar SC, Kruger WD, Testa JR (June 2004). "Growth inhibition and induction of apoptosis in mesothelioma cells by selenium and dependence on selenoprotein SEP15 genotype". Oncogene. 23 (29): 5032–40. doi:10.1038/sj.onc.1207683. PMID 15107826.
- Wellenreuther R, Schupp I, Poustka A, Wiemann S (June 2004). "SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones". BMC Genomics. 5 (1): 36. doi:10.1186/1471-2164-5-36. PMC 436056. PMID 15198809.