SAM riboswitch (S-box leader)
The SAM riboswitch (also known as the S-box leader and the SAM-I riboswitch) is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form.[1][2][3] When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase read-through the downstream gene.[4] When the SAM is bound to the aptamer, the anti-terminator is sequestered by an anti-anti-terminator; the terminator forms and terminates the transcription.[4][5] However, many SAM riboswitches are likely to regulate gene expression at the level of translation.
| SAM riboswitch (S-box leader) | |
|---|---|
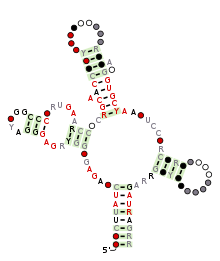 Predicted secondary structure and sequence conservation of SAM | |
| Identifiers | |
| Symbol | SAM |
| Alt. Symbols | S_box |
| Rfam | RF00162 CL00012 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Bacteria |
| SO | 0000035 |
| PDB structures | PDBe |
Structure organization
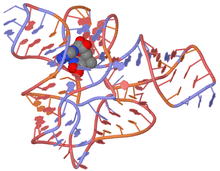
The structure of the SAM riboswitch has been determined with X-ray crystallography.[6] The SAM riboswitch is organized about a four way junction, with two sets of coaxially stacked helices arranged side-by-side. These stacks are held together by a pseudoknot formed between the loop on the end of stem P2 and the J3/4 joining region. The formation of the pseudoknot is facilitated by a protein-independent kink turn that induces a 100° bend into P2. Ribosomal proteins, known to bind kink-turns in the ribosome, favor SAM aptamer folding by interacting with P2 kink-turn motif.[7] Both the kink-turn and the pseudoknot are critical to the establishment of the global fold and productive binding. The binding pocket is split between conserved, tandem AU pairs in stem P1, the conserved G in the J1/2 joining region, and the conserved asymmetric bulge in stem P3. The adenosyl and methionine main-chain moieties of S-Adenosyl methionine (SAM) are recognized through hydrogen-bonding into the bulge in P3 and the conserved G in J1/2. The methyl group is recognized indirectly through the charged sulfur, which forms an electrostatic interaction with the negative surface potential created by the tandem AU pairs in the minor groove of P1. These pairs are highly conserved and alterations to the orientation of these pairs, as well the identity of the bases in the pairs (i.e., GC pairs instead of AU pairs) result in reduced affinity for SAM. Affinity for SAH, however, is unaffected by changes to the P1 sequence, further supporting the idea that the interaction between SAM and the P1 helix is electrostatic in nature.
Other structural classes of SAM-binding riboswitches have been found that are entirely different from that of the SAM-I riboswitch. These unrelated SAM-binding riboswitches are SAM-II riboswitches, SAM-III riboswitches and SAM–SAH riboswitches. The details of these riboswitches are covered in their articles.
There are also classes of SAM-binding riboswitches that are structurally related to SAM-I riboswitches. SAM-IV riboswitches, appears to share a similar ligand-binding site with that of SAM-I riboswitches, but in the context of a discreet distinct scaffold. In SAM-IV, the P4 stem is located downstream of the P1 stem and interacts, by a second pseudoknot, with P3, while the kink turn is absent. SAM-I/IV variant riboswitches[8] combine features of SAM-I and SAM-IV riboswitches. For example, they have the P4 outside of the multistem junction, like SAM-IV riboswitches, but also often have a SAM-I–like P4 within the multistem junction. However, unlike SAM-I and SAM-IV riboswitches, the variant riboswitches entirely lack an internal loop in P2. As noted above, this internal loop in SAM-I riboswitches forms a kink turn motif that allows the RNA to form a pseudoknot. The variant riboswitches lack an internal loop that might allow such a turn, and also show no signs of forming the SAM-I–like pseudoknot.
References
- Grundy FJ, Henkin TM (1998). "The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria". Mol. Microbiol. 30 (4): 737–749. doi:10.1046/j.1365-2958.1998.01105.x. PMID 10094622.
- Epshtein, V; Mironov AS; Nudler E (2003). "The riboswitch-mediated control of sulfur metabolism in bacteria". Proc Natl Acad Sci USA. 100 (9): 5052–5056. doi:10.1073/pnas.0531307100. PMC 154296. PMID 12702767.
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR (2003). "An mRNA structure that controls gene expression by binding S-adenosylmethionine". Nat. Struct. Biol. 10 (9): 701–707. doi:10.1038/nsb967. PMID 12910260.
- Winkler, W., Nahvi, A., Sudarsan, N., Barrick, J., and Breaker, R. (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Structural Biology, 10(9), 701–707.
- Epshtein, V., Mironov, A., and Nudler, E. (2003) The riboswitch-mediated control of sulfur metabolism in bacteria. Proceedings of the National Academy of Sciences of the USA, 100, 5052–5056.
- Montange, RK; Batey RT (2006). "Structure of the S-adenosylmethionine riboswitch regulatory mRNA element". Nature. 441 (7097): 1172–1175. doi:10.1038/nature04819. PMID 16810258.
- Heppell, B; Lafontaine DA (2008). "Folding of the SAM aptamer is determined by the formation of a K-turn-dependant pseudoknot". Biochemistry. 47 (6): 1490–1499. doi:10.1021/bi701164y. PMID 18205390.
- Weinberg Z, Wang JX, Bogue J, et al. (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol. 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571. PMID 20230605.
External links
- Page for SAM riboswitch (S box leader) at Rfam (a Wikipedia clone)
- PDB entry for the SAM riboswitch tertiary structure