Q-system (genetics)
Q-system is a genetic tool that allows to express transgenes in a living organism.[1] Originally the Q-system was developed [2][3] for use in the vinegar fly Drosophila melanogaster, and was rapidly adapted for use in cultured mammalian cells,[2] zebrafish,[4] worms[5] and mosquitoes.[6] The Q-system utilizes genes from the qa cluster[7] of the bread fungus Neurospora crassa, and consists of four components: the transcriptional activator (QF/QF2/QF2w), the enhancer QUAS, the repressor QS, and the chemical de-repressor quinic acid. Similarly to GAL4/UAS[8] and LexA/LexAop,[9] the Q-system is a binary expression system that allows to express reporters or effectors (e.g. fluorescent proteins, ion channels, toxins and other genes) in a defined subpopulation of cells with the purpose of visualising these cells or altering their function. In addition, GAL4/UAS, LexA/LexAop and the Q-system function independently of each other and can be used simultaneously to achieve a desired pattern of reporter expression, or to express several reporters in different subsets of cells.
Origin
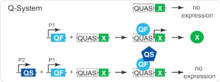
The Q-system is based on two out of the seven genes of the qa gene cluster of the bread fungus Neurospora crassa.[7] The genes of the qa cluster are responsible for the catabolism of quinic acid, which is used by the fungus as a carbon source in conditions of low glucose.[7] The cluster contains a transcriptional activator qa-1F, a transcriptional repressor qa-1S, and five structural genes. The qa-1F binds to a specific DNA sequence, found upstream of the qa genes. The presence of quinic acid disrupts interaction between qa-1F and qa-1S, thus disinhibiting the transcriptional activity of qa-1F. Genes qa-1F, qa-1S and the DNA binding sequence of qa-1F form the basis of the Q-system. The genes were renamed to simplify their use as follows: transcriptional activator qa-1F as QF, repressor qa-1S as QS, and the DNA binding sequence as QUAS.[2] The quinic acid represents the fourth component of the Q-system. The original transactivator QF appeared to be toxic when expressed broadly in Drosophila. To overcome this problem, two new transactivators were developed: QF2 and QF2w[3]
Use in Drosophila
Basic use
The Q-system functions similarly to, and independently of, the GAL4/UAS[8] and the LexA/LexAop [9] systems. QF, QF2 and QF2w are analogous to GAL4 and LexA, and their expression is usually under the control of cell-type specific promoter, such as nsyb (to target neurons) or tubulin (to target all cells). QUAS is analogous to UAS and LexAop, and is placed upstream of an effector gene, such as GFP. QS is analogous to GAL80, and may be driven by any promoter (e.g. tubulin-QS). Quinic acid is a unique feature of the Q-system, and it must be fed to the flies or maggots in order to alleviate the QS-induced repression. In some ways, quinic acid is analogous to temperature in the case of GAL80ts. In its basic form, two transgenic fly lines, one containing a QF transgene and the other one containing a QUAS transgene, are crossed together. Their progeny that had both a QF transgene and a QUAS transgene will be expressing a reporter gene in a subset of cells (e.g. nsyb-QF2, QUAS-GFP flies express GFP in all neurons). If a fly also expresses QS in some of the cells, the activity of QF will be repressed in these cells, but it may be restored of a fly is fed quinic acid (e.g. a nsyb-QF2, QUAS-GFP, tub-QS fly expresses no GFP when its diet doesn't contain quinic acid, and expresses GFP in its neurons when fed quinic acid).[2][3] The use of QS repressor and quinic acid allows to fine-tune the temporal control of transgene expression.
Chimeric transactivators
Chimeric transactivators GAL4QF[3] and LexAQF[3] allow to combine the use of all three binary expression systems. GAL4QF binds to UAS, and may be repressed by QS while being unaffected by GAL80. Similarly, LexAQF binds to LexAop, and may be repressed by QS. LexAQF represents a useful extension of the LexA/LexAop system that doesn't have its own repressor.
Intersectional expression
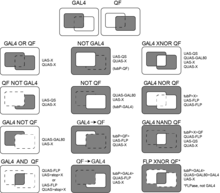
A variety of expression patterns may be achieved by combination of the three binary expression systems and the FLP/FRT or other recombinases.[10] Expression patterns may be constructed as AND, OR, NOR etc. logic gates [1][2] to e.g. narrow down expression patterns of available GAL4 lines. The resulting expression pattern somewhat depends on the developmental timing of activation of the transcription factors (discussed in [1]).
Use in other organisms
Q-system appeared to be working successfully in a variety of organisms. It has been used to drive expression of luciferase, as a proof of principle, in cultured mammalian cells.[2] In zebrafish[4] the Q-system has been successfully used with several tissue-specific promoters, and was shown to work independently of the GAL4/UAS system when expressed in the same cell. In C. elegans[5] the Q-system has been shown to work in muscles and in neuronal tissue. In 2016, the Q-system was used to target, for the first time, the olfactory neurons of malaria mosquitoes Anopheles gambiae.[6] In 2019, the Q-system in Anopheles mosquitoes was used to examine the functional responses of olfactory neurons to odors.[11] In 2019, the Q-system was introduced into the Aedes aegypti mosquito to capture tissue specific expression patterns.[12] These successes make the Q-system the system of choice when developing genetic tools for other organisms. Currently the main shortcoming of the Q-system is the low number of available transgenic lines, but it will be overcome as the scientific community creates and shares these resources, such as by the use of the GAL4>QF2 HACK system to convert existing GAL4 transgenic insertions to QF2.[13] DNA binding domain of QF2 fused with VP16 transcriptional activator domain was successfully applied in Penicillium to gain control over the penicillin producing secondary metabolite gene cluster in a scalable manner. [14]
References
- Riabinina O, Potter CJ (2016). "The Q-System: A Versatile Expression System for Drosophila". Methods in Molecular Biology. 1478: 53–78. doi:10.1007/978-1-4939-6371-3_3. ISBN 978-1-4939-6369-0. PMC 5270762. PMID 27730575.
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L (April 2010). "The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis". Cell. 141 (3): 536–48. doi:10.1016/j.cell.2010.02.025. PMC 2883883. PMID 20434990.
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ (March 2015). "Improved and expanded Q-system reagents for genetic manipulations". Nature Methods. 12 (3): 219–22, 5 p following 222. doi:10.1038/nmeth.3250. PMC 4344399. PMID 25581800.
- Subedi A, Macurak M, Gee ST, Monge E, Goll MG, Potter CJ, Parsons MJ, Halpern ME (April 2014). "Adoption of the Q transcriptional regulatory system for zebrafish transgenesis". Methods. 66 (3): 433–40. doi:10.1016/j.ymeth.2013.06.012. PMC 3883888. PMID 23792917.
- Wei X, Potter CJ, Luo L, Shen K (March 2012). "Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans". Nature Methods. 9 (4): 391–5. doi:10.1038/nmeth.1929. PMC 3846601. PMID 22406855.
- Riabinina O, Task D, Marr E, Lin CC, Alford R, O'Brochta DA, Potter CJ (October 2016). "Organization of olfactory centres in the malaria mosquito Anopheles gambiae". Nature Communications. 7: 13010. Bibcode:2016NatCo...713010R. doi:10.1038/ncomms13010. PMC 5063964. PMID 27694947.
- Giles NH, Geever RF, Asch DK, Avalos J, Case ME (1991). "The Wilhelmine E. Key 1989 invitational lecture. Organization and regulation of the qa (quinic acid) genes in Neurospora crassa and other fungi". The Journal of Heredity. 82 (1): 1–7. doi:10.1093/jhered/82.1.1. PMID 1825499.
- Brand AH, Perrimon N (June 1993). "Targeted gene expression as a means of altering cell fates and generating dominant phenotypes". Development. 118 (2): 401–15. PMID 8223268.
- Lai SL, Lee T (May 2006). "Genetic mosaic with dual binary transcriptional systems in Drosophila". Nature Neuroscience. 9 (5): 703–9. doi:10.1038/nn1681. PMID 16582903.
- Bischof J, Basler K (2008). "Recombinases and their use in gene activation, gene inactivation, and transgenesis". Methods in Molecular Biology. 420: 175–95. doi:10.1007/978-1-59745-583-1_10. ISBN 978-1-58829-817-1. PMID 18641947.
- Afify A, Betz JF, Riabinina O, Lahondère C, Potter CJ (October 2019). "Commonly Used Insect Repellents Hide Human Odors from Anopheles Mosquitoes". Current Biology. 0: 3669–3680.e5. doi:10.1016/j.cub.2019.09.007. PMC 6832857. PMID 31630950.
- Matthews BJ, Younger MA, Vosshall LB (May 2019). "Aedes aegypti". eLife. 8: e43963. doi:10.7554/eLife.43963. PMC 6597239. PMID 31112133.
- Lin CC, Potter CJ (August 2016). "Editing Transgenic DNA Components by Inducible Gene Replacement in Drosophila melanogaster". Genetics. 203 (4): 1613–28. doi:10.1534/genetics.116.191783. PMC 4981265. PMID 27334272.
- Mózsik L, et al. (November 2019). "Synthetic control devices for gene regulation in Penicillium chrysogenum". Microb Cell Fact. 18 (203). doi:10.1186/s12934-019-1253-3.