petite mutation
petite (ρ–) is a mutant first discovered in the yeast Saccharomyces cerevisiae. Due to the defect in the respiratory chain, 'petite' yeast are unable to grow on media containing only non-fermentable carbon sources (such as glycerol or ethanol) and form small colonies when grown in the presence of fermentable carbon sources (such as glucose). The petite phenotype can be caused by the absence of, or mutations in, mitochondrial DNA (termed "cytoplasmic Petites"), or by mutations in nuclear-encoded genes involved in oxidative phosphorylation.[1][2] A neutral petite produces all wild type progeny when crossed with wild type.
petite mutations can be induced using a variety of mutagens, including DNA intercalating agents, as well as chemicals that can interfere with DNA synthesis in growing cells.[2] Mutagens that create Petites are implicated in increased rates of degenerative diseases and in the aging process.
Overview
A mutation that produces small (petite" > petite) anaerobic-like colonies had shown first in Yeast Saccharomyces cerevisiae and described by Boris Ephrussi and his co-workers in 1949 in Gif-sur-Yvette, France.[3] The cells of petite colonies were smaller than those of wild-type colonies, but the term “petite” refers only to colony size and not the individual cell size.[3]
History
Over 50 years ago, in a lab in France, Ephrussi, et al. discovered a non-Mendelian inherited factor that is essential to respiration in the yeast, Saccharomyces cerevisiae. S. cerevisiae without this factor, known as the ρ-factor, is described by the development of small colonies when compared to the wild-type yeast.[4] These smaller colonies were dubbed petite colonies. These petite mutants were observed to be spontaneously produced naturally at a rate of 0.1%-1.0% every generation.[4][5] They also found that treatment of wild-type S. cerevisiae with DNA-intercalating agents would more rapidly produce this mutation.[4]
Schatz identified a region of the yeast’s nuclear DNA that was associated with the mitochondria in 1964. Later, it was discovered that mutants without the ρ-factor had no mitochondrial DNA (called ρ0 isolates), or that they possessed a difference in density or amount of the mitochondrial DNA (called ρ− isolates). The use of electron microscopy to view the DNA in the mitochondrial matrix helped to verify the actuality of the mitochondrial genome.[4][5][6]
S. cerevisiae has since become a useful model for aging. It has been shown that as yeast ages, it loses functional mitochondrial DNA, which leads to replicative senescence, or the inability to further replicate.[4] It has been suggested that there is a link between mitochondrial DNA loss and replicative life span (RLS), or the number of times a cell can reproduce before it dies, as it has been found that an increase in RLS is established with the same changes in the genome that enhance the propagation of cells that do not contain mitochondrial DNA. Genetic screens for replicative life span associated genes and pathways could be made easier and quicker by selecting genetic suppressors of the petite negative mutants.[4]
Causes
The petite is characterized by a deficiency in cytochromes (a, a3 + b) and a lack of respiratory enzymes which engage in respiration in mitochondria.[7] Due to the error in the respiratory chain pathway, 'petite' yeast is incapable of growing on media containing only non-fermentable carbon sources (such as glycerol or ethanol) and forming small colonies when grown in the presence of fermentable carbon sources (such as glucose).[8] The absence of mitochondria can cause the petite phenotype, or by deletion mutations in mitochondrial DNA (termed "cytoplasmic Petites") which is a deletion mutation, or by mutations in nuclear-encoded genes involved in oxidative phosphorylation.
Experiment
Petite mutants can be generated in the laboratory by using high-efficiency treatments such as acriflavine, ethidium bromide, and other intercalating agents.[9] Their mechanisms work to break down and cause the eventual loss of mitochondrial DNA: if the treatment time increases, the amount of mitochondrial DNA will decrease. After prolonged treatment, petites containing no detectable mitochondrial DNA were obtained.[7] It is useful approach to illustrate the function of mitochondrial DNA in yeast growth.
Petite mutation inheritance
The inheritance pattern of genes existing in the cell organelles such as mitochondria which named cytoplasmic inheritance differs from nuclear genes pattern.
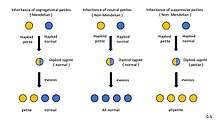
Petite mutation inheritance types
Petite mutants show extranuclear inheritance.The inheritance pattern varying with the type of petite involved.
Segregational petites (pet–): mutants are created by nuclear mutations and exhibit Mendelian 1:1 segregation.[9]
Neutral petites (rho–N): Neutral petite when crossed to wild-type, all offspring are wild-type. It has inherited normal mitochondrial DNA from wild-type parent, which is replicated in the offspring.[3]
Suppressive petites (rho–S): crosses between petite and wild-type, all offspring are petite, showing "dominant" behavior to suppress wild-type mitochondrial function.[3]
Most petite mutants of S. cerevisiae are of a suppressive type, and they differ from neutral petite by affecting the wild-type, although both are a mutation in mitochondrial DNA. Mitochondrial genome of yeast will be the first eukaryotic genome to be understood in terms of both structure and function and this should smooth the way to understand the evolution of organelle genomes and its relationship with nuclear genomes.It is evident that Ephrussi’s work not only opened the field of extrachromosomal genetics, but also provide a fantastic incentive for the investigations which followed up to this day.[3]
Though S. cerevisiae has been extensively studied in this and other areas, it is difficult to say if the molecular mechanisms of this process in the mitochondrial DNA are conserved across other yeast species. Other yeast species, such as Kluyveromyces lactis, Saccharomyces castellii, and Candida albicans have all shown to produce petite negative mutants. Potentially, these yeasts have a different inheritance system in place for their mitochondrial genome than S. cerevisiae does.[4][5]
The frequency at which S. castellii spontaneously produces petites is similar to that of S. cerevisiae, with the mitochondrial DNA of those petites being highly altered via deletion and rearrangement. Suppressive petites of S. cerevisiae are the most commonly observed spontaneously created mutants, whereas in S. castellii, the most commonly observed spontaneous mutant is the neutral petite, further leading to speculation that the transference of this mutation differs between species.[1][4][5]
References
- Lipinski, Kamil A.; Kaniak-Golik, Aneta; Golik, Pawel (June 2010). "Maintenance and expression of the S. cerevisiae mitochondrial genome—From genetics to evolution and systems biology". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1797 (6–7): 1086–1098. doi:10.1016/j.bbabio.2009.12.019. ISSN 0005-2728. PMID 20056105.
- Ferguson, L. R.; von Borstel, R. C. (1992-01-01). "Induction of the cytoplasmic 'petite' mutation by chemical and physical agents in Saccharomyces cerevisiae". Mutation Research. 265 (1): 103–148. doi:10.1016/0027-5107(92)90042-Z. ISSN 0027-5107. PMID 1370239.
- Bernardi, Giorgio (1979-09-01). "The petite mutation in yeast". Trends in Biochemical Sciences. 4 (9): 197–201. doi:10.1016/0968-0004(79)90079-3.
- Dunn, Cory D. (2011-08-09). "Running on empty: Does mitochondrial DNA mutation limit replicative lifespan in yeast?". BioEssays. 33 (10): 742–748. doi:10.1002/bies.201100050. ISSN 0265-9247. PMID 21826691.
- Kochmak, S. A.; Knorre, D. A.; Sokolov, S. S.; Severin, F. F. (2011-02-01). "Physiological scenarios of programmed loss of mitochondrial DNA function and death of yeast". Biochemistry (Moscow). 76 (2): 167–171. doi:10.1134/s0006297911020015. ISSN 0006-2979. PMID 21568848.
- Lipinski, Kamil A.; Kaniak-Golik, Aneta; Golik, Pawel (2010-06-01). "Maintenance and expression of the S. cerevisiae mitochondrial genome—From genetics to evolution and systems biology". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1797 (6–7): 1086–1098. doi:10.1016/j.bbabio.2009.12.019. ISSN 0005-2728. PMID 20056105.
- Goldring, Elizabeth S.; Grossman, Lawrence I.; Marmur, Julius (July 1971). "Petite Mutation in Yeast". Journal of Bacteriology. 107 (1): 377–381. doi:10.1128/JB.107.1.377-381.1971. ISSN 0021-9193. PMC 246929. PMID 5563875.
- Heslot, H.; Goffeau, A.; Louis, C. (1970-10-01). "Respiratory Metabolism of a "Petite Negative" Yeast Schizosaccharomyces pombe 972h−". Journal of Bacteriology. 104 (1): 473–481. doi:10.1128/JB.104.1.473-481.1970. ISSN 0021-9193. PMC 248232. PMID 4394400.
- Heslot, H.; Louis, C.; Goffeau, A. (1970-10-01). "Segregational Respiratory-Deficient Mutants of a "Petite Negative" Yeast Schizosaccharomyces pombe 972h−". Journal of Bacteriology. 104 (1): 482–491. doi:10.1128/JB.104.1.482-491.1970. ISSN 0021-9193. PMC 248233. PMID 5473904.