Neofunctionalization
Neofunctionalization, one of the possible outcomes of functional divergence, occurs when one gene copy, or paralog, takes on a totally new function after a gene duplication event. Neofunctionalization is an adaptive mutation process; meaning one of the gene copies must mutate to develop a function that was not present in the ancestral gene.[1][2][3] In other words, one of the duplicates retains its original function, while the other accumulates molecular changes such that, in time, it can perform a different task.[4] This process is thought to be free of selective pressure because one gene copy can mutate without adversely affecting the fitness of the organism since ancestral function is retained in the other copy.[5][6][7][8]
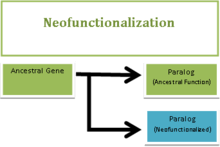
The process
The process of Neofunctionalization begins with a gene duplication event, which is thought to occur as a defense mechanism against the accumulation of deleterious mutations.[6][8][9] Following the gene duplication event there are two identical copies of the ancestral gene performing exactly the same function. This redundancy allows one the copies to take on a new function. In the event that the new function is advantageous, natural selection positively selects for it and the new mutation becomes fixed in the population.[3][10] The occurrence of Neofunctionalization can most often be attributed to changes in the coding region or changes in the regulatory elements of a gene.[8] It is much more rare to see major changes in protein function, such as subunit structure or substrate and ligand affinity, as a result of Neofunctionalization.[8]
Selective constraints
Neofunctionalization is also commonly referred to as "mutation during non-functionality” or “mutation during redundancy”.[11] Regardless of if the mutation arises after non-functionality of a gene or due to redundant gene copies, the important aspect is that in both scenarios one copy of the duplicated gene is freed from selective constraints and by chance acquires a new function which is then improved by natural selection.[8] This process is thought to occur very rarely in evolution for two major reasons. The first reason is that functional changes typically require a large number of amino acid changes; which has a low probability of occurrence. Secondly, because deleterious mutations occur much more frequently than advantageous mutations in evolution.[8] This makes the likelihood that gene function is lost over time (i.e. pseudogenization) far greater than the likelihood of the emergence of a new gene function.[10] Walsh discovered that the relative probability of Neofunctionalization is determined by the selective advantage and the relative rate of advantageous mutations.[12] This was proven in his derivation of the relative probability of Neofunctionalization to pseudogenization, which is given by: where ρ is the ratio of advantageous mutation rate to null mutation rate and S is the population selection 4NeS (Ne: effective population size S: selection intensity).[12]
Classical model
In 1936, Muller originally proposed Neofunctionalization as a possible outcome of a gene duplication event.[13] In 1970, Ohno suggested that Neofunctionalization was the only evolutionary mechanism that gave rise to new gene functions in a population.[8] He also believed that Neofunctionalization was the only alternative to pseudogenization.[2] Ohta (1987) was among the first to suggest that other mechanisms may exist for the preservation of duplicated genes in the population.[8] Today, subfunctionalization is a widely accepted alternative fixation process for gene duplicates in the population and is currently the only other possible outcome of functional divergence.[2]
Neosubfunctionalization
Neosubfunctionalization occurs when Neofunctionalization is the end result of subfunctionalization. In other words, once a gene duplication event occurs forming parologs that after an evolutionary period subfunctionalize, one gene copy continues on this evolutionary journey and accumulates mutations that give rise to a new function.[8][14] Some believe that Neofunctionalization is the end stage for all subfunctionalized genes. For instance, according to Rastogi and Liberles “Neofunctionalization is the terminal fate of all duplicate gene copies retained in the genome and subfuctionlization merely exist as a transient state to preserve the duplicate gene copy.”[2] The results of their study become punctuated as population size increases.
Examples
The evolution of the antifreeze protein in the Antarctic zoarcid fish L. dearborni provides a prime example of Neofunctionalization after gene duplication. In the case of the Antarctic zoarcid fish type III antifreeze protein gene (AFPIII; P12102) diverged from a paralogous copy of sialic acid synthase (SAS) gene.[15] The ancestral SAS gene was found to have both sialic acid synthase and rudimentary ice-binding functionalities. After duplication one of the paralogs began to accumulate mutations that lead to the replacement of SAS domains of the gene allowing for further development and optimization of the antifreeze functionality.[15] The new gene is now capable of noncolligative freezing-point depression, and thus is neofunctionalized.[15] This specialization allows Antarctic zoarcid fish to survive in the frigid temperatures of the Antarctic Seas.
Model limitations
Limitations exist in Neofunctionalization as model for functional divergence primarily because:
- the amount of nucleotide changes giving rise to a new function must be very minimal; making the probability for pseudogenization much higher than neofunctionalization after a gene duplication event.[8]
- After a gene duplication event both copies may be subjected to selective pressure equivalent to that constraining the ancestral gene; meaning that neither copy is available for Neofunctionalization.[8]
- In many cases positive Darwinian selection presents a more parsimonious explanation for the divergence of multigene families.[8]
See also
References
- Kleinjan, Dirk A.; Bancewicz, Ruth M.; Gautier, Philippe; Dahm, Ralf; Schonthaler, Helia B.; Damante, Giuseppe; Seawright, Anne; Hever, Ann M.; Yeyati, Patricia L.; van Heyningen, Veronica; Coutinho, Pedro (1 January 2008). "Subfunctionalization of Duplicated Zebrafish pax6 Genes by cis-Regulatory Divergence". PLoS Genetics. 4 (2): e29. doi:10.1371/journal.pgen.0040029. PMC 2242813. PMID 18282108.
- Rastogi, S.; Liberles, D. A. (2005). "Subfunctionalization of duplicated genes as a transition state to neofunctionalization". BMC Evolutionary Biology. 5 (1): 28. doi:10.1186/1471-2148-5-28. PMC 1112588. PMID 15831095.
- Conrad, B.; Antonarakis, S. E. (2007). "Gene duplication: a drive for phenotypic diversity and cause of human disease". Annual Review of Genomics and Human Genetics. 8: 17–35. doi:10.1146/annurev.genom.8.021307.110233. PMID 17386002.
- S. Ohno, Evolution by Gene Duplication. New York, Heidelberg, Berlin: Springer-Verlag, 1970, pp. 59-87
- Sémon, M.; Wolfe, K. H. (2008). "Preferential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis". Proceedings of the National Academy of Sciences of the United States of America. 105 (24): 8333–8. Bibcode:2008PNAS..105.8333S. doi:10.1073/pnas.0708705105. PMC 2448837. PMID 18541921.
- R. De Smet and Y. Van de Peer, “Redundancy and rewiring of genetic networks following genome-wide duplication events.,” Current Opinion in Plant Biology, pp. 1-9, Feb. 2012
- Ruby, J. G.; Stark, A.; Johnston, W. K.; Kellis, M.; Bartel, D. P.; Lai, E. C. (2007). "Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs". Genome Research. 17 (12): 1850–64. doi:10.1101/gr.6597907. PMC 2099593. PMID 17989254.
- D. Graur and W.-H. Li, Fundamentals of Molecular Evolution, Second Ed. Sinauer Associates, Inc., 2000.
- G. D. Amoutzias, Y. He, J. Gordon, D. Mossialos, S. G. Oliver, and Y. Van de Peer, “Posttranslational regulation impacts the fate of duplicated genes.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 107, no. 7, pp. 2967-71, Feb. 2010
- H. Innan, “Population genetic models of duplicated genes.,” Genetica, vol. 137, no. 1, pp. 19-37, Sep. 2009
- A. Hughes, Adaptive Evolution of Genes and Genomes. New York: Oxford University Press, 1999
- Lynch, Michael; Force, Allan (2000-01-01). "The Probability of Duplicate Gene Preservation by Subfunctionalization". Genetics. 154 (1): 459–473. ISSN 0016-6731. PMC 1460895. PMID 10629003.
- Muller, Hermann J. (1936). "Bar duplication". Science. 83 (2161): 528–530. Bibcode:1936Sci....83..528M. doi:10.1126/science.83.2161.528-a. PMID 17806465.
- X. He and J. Zhang, “Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution.,” Genetics, vol. 169, no. 2, pp. 1157-1164, 2005
- Deng, C.; Cheng, C.- H. C.; Ye, H.; He, X.; Chen, L. (29 November 2010). "Evolution of an antifreeze protein by neofunctionalization under escape from adaptive conflict". Proceedings of the National Academy of Sciences. 107 (50): 21593–21598. Bibcode:2010PNAS..10721593D. doi:10.1073/pnas.1007883107. PMC 3003108. PMID 21115821.