Mitotic recombination
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual.[1] Additionally, mitotic recombination can result in the expression of recessive genes in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive genes.[1][2] Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication (but prior to cell division). Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.[3]
Discovery
The discovery of mitotic recombination came from the observation of twin spotting in Drosophila melanogaster. This twin spotting, or mosaic spotting, was observed in D. melanogaster as early as 1925, but it was only in 1936 that Curt Stern explained it as a result of mitotic recombination. Prior to Stern's work, it was hypothesized that twin spotting happened because certain genes had the ability to eliminate the chromosome on which they were located.[4] Later experiments uncovered when mitotic recombination occurs in the cell cycle and the mechanisms behind recombination.
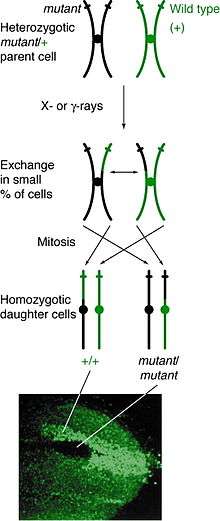
Occurrence
Mitotic recombination can happen at any locus but is observable in individuals that are heterozygous at a given locus. If a crossover event between non-sister chromatids affects that locus, then both homologous chromosomes will have one chromatid containing each genotype. The resulting phenotype of the daughter cells depends on how the chromosomes line up on the metaphase plate. If the chromatids containing different alleles line up on the same side of the plate, then the resulting daughter cells will appear heterozygous and be undetectable, despite the crossover event. However, if chromatids containing the same alleles line up on the same side, the daughter cells will be homozygous at that locus. This results in twin spotting, where one cell presents the homozygous recessive phenotype and the other cell has the homozygous wild type phenotype. If those daughter cells go on to replicate and divide, the twin spots will continue to grow and reflect the differential phenotype.
Mitotic recombination takes place during interphase. It has been suggested that recombination takes place during G1, when the DNA is in its 2-strand phase, and replicated during DNA synthesis.[5] It is also possible to have the DNA break leading to mitotic recombination happen during G1, but for the repair to happen after replication.[6][7]
Response to DNA damage
In the budding yeast Saccharomyces cerevisiae, mutations in several genes needed for mitotic (and meiotic) recombination cause increased sensitivity to inactivation by radiation and/or genotoxic chemicals.[8] For example, gene rad52 is required for mitotic recombination[9] as well as meiotic recombination.[10] Rad52 mutant yeast cells have increased sensitivity to killing by X-rays, methyl methanesulfonate and the DNA crosslinking agent 8-methoxypsoralen-plus-UV light, suggesting that mitotic recombinational repair is required for removal of the different DNA damages caused by these agents.
Mechanisms
The mechanisms behind mitotic recombination are similar to those behind meiotic recombination. These include sister chromatid exchange and mechanisms related to DNA double strand break repair by homologous recombination such as single-strand annealing, synthesis-dependent strand annealing (SDSA), and gene conversion through a double-Holliday Junction intermediate or SDSA. In addition, non-homologous mitotic recombination is a possibility and can often be attributed to non-homologous end joining.[6][7][11][12]
Method
There are several theories on how mitotic crossover occurs. In the simple crossover model, the two homologous chromosomes overlap on or near a common Chromosomal fragile site (CFS). This leads to a double-strand break,[13] which is then repaired using one of the two strands. This can lead to the two chromatids switching places. In another model, two overlapping sister chromatids form a double Holliday junction at a common repeat site and are later sheared in such a way that they switch places. In either model, the chromosomes are not guaranteed to trade evenly, or even to rejoin on opposite sides thus most patterns of cleavage do not result in any crossover event. Uneven trading introduces many of the deleterious effects of mitotic crossover.
Alternatively, a crossover can occur during DNA repair[14] if, due to extensive damage, the homologous chromosome is chosen to be the template over the sister chromatid. This leads to gene synthesis since one copy of the allele is copied across from the homologous chromosome and then synthesized into the breach on the damaged chromosome. The net effect of this would be one heterozygous chromosome and one homozygous chromosome.
Advantages and disadvantages
Mitotic crossover is known to occur in D. melanogaster, some asexually reproducing fungi and in normal human cells, where the event may allow normally recessive cancer-causing genes to be expressed and thus predispose the cell in which it occurs to the development of cancer. Alternately, a cell may become a homozygous mutant for a tumor-suppressing gene, leading to the same result.[2] For example, Bloom's syndrome is caused by a mutation in RecQ helicase, which plays a role in DNA replication and repair. This mutation leads to high rates of mitotic recombination in mice, and this recombination rate is in turn responsible for causing tumor susceptibility in those mice.[15] At the same time, mitotic recombination may be beneficial: it may play an important role in repairing double stranded breaks, and it may be beneficial to the organism if having homozygous dominant alleles is more functional than the heterozygous state.[2] For use in experimentation with genomes in model organisms such as Drosophila melanogaster, mitotic recombination can be induced via X-ray and the FLP-FRT recombination system.[16]
References
- Hartl, Daniel L. and Maryellen Ruvolo (2012). Genetics: Analysis of Genetics and Genomes. Burlington: Jones & Bartlett.
- Tischfield, Jay A. (November 1997). "Loss of Heterozygosity, or: How I learned to Stop Worrying and Love Mitotic Recombination". American Journal of Human Genetics. 61 (5): 995–999. doi:10.1086/301617. PMC 1716040. PMID 9345110.
- Moynahan ME, Jasin M (2010). "Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis". Nat. Rev. Mol. Cell Biol. 11 (3): 196–207. doi:10.1038/nrm2851. PMC 3261768. PMID 20177395.
- Stern, Curt (1936). "Somatic Crossing Over and Segregation in Drosophila Melanogaster". Genetics. 21 (6): 625–730.
- Esposito, Michael S (September 1978). "Evidence that Spontaneous Mitotic Recombination Occurs at the Two-Strand Stage". Proceedings of the National Academy of Sciences of the USA. 75 (9): 4436–4440. doi:10.1073/pnas.75.9.4436. PMC 336130. PMID 360220.
- Lee, Phoebe S.; Greenwell, Patricia W.; Dominska, Margaret; Gawel, Malgorzata; Hamilton, Monica; Petes, Thomas D. (2009). "A Fine-Structure Map of Spontaneous Mitotic Crossovers in the Yeast Saccharomyces cerevisiae". PLoS Genet. 5 (3): e1000410. doi:10.1371/journal.pgen.1000410. PMC 2646836. PMID 19282969.
- LaFave, MC; J Sekelsky (2009). "Mitotic Recombination: Why? When? How? Where?". PLoS Genet. 5 (3): e1000411. doi:10.1371/journal.pgen.1000411. PMC 2648873. PMID 19282976.
- Haynes, R.H. & Kunz, B.A. (1981). DNA repair and mutagenesis in yeast. In: Strathern, J; Jones, E; Broach J. editors. The Molecular Biology of the Yeast Saccharomyces. Life Cycle and Inheritance. Cold Spring Harbor, N.Y., Cold Spring Harbor Laboratory, 371-414.
- Malone RE, Esposito RE (1980). "The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast". Proc. Natl. Acad. Sci. U.S.A. 77 (1): 503–7. doi:10.1073/pnas.77.1.503. PMC 348300. PMID 6987653.
- Game JC, Zamb TJ, Braun RJ, Resnick M, Roth RM (1980). "The Role of Radiation (rad) Genes in Meiotic Recombination in Yeast". Genetics. 94 (1): 51–68. PMC 1214137. PMID 17248996.
- Helleday, Thomas (2003). "Pathways for Mitotic Homologous Recombination in Mammalian Cells". Mutation Research. 532 (1–2): 103–115. doi:10.1016/j.mrfmmm.2003.08.013.
- Pâques, Frédéric; James E. Haber (1999). "Multiple Pathways of Recombination Induced by Double-Strand Breaks in Saccharomyces cerevisiae". Microbiology and Molecular Biology Reviews. 63 (2): 349–404. PMC 98970. PMID 10357855.
- Helleday, T. "Double-Strand Break Repair via Double Holliday Junctions (Szostak Model)". Animation. MIT.
- Helleday, Thomas (27 November 2003). "Pathways for mitotic homologous recombination in mammalian cells" (PDF). Mutation Research. 532 (1–2): 103–115. doi:10.1016/j.mrfmmm.2003.08.013. Archived from the original (PDF) on 24 May 2014. Retrieved 2012-12-26.
- Luo, Guangbin; et al. (2000). "Cancer predisposition caused by elevated mitotic recombination in Bloom mice". Nature Genetics. 26: 424–429. doi:10.1038/82548. PMID 11101838.
- Xu, T; GM Rubin (April 1993). "Analysis of genetic mosaics in developing and adult Drosophila tissues". Development. 117 (4): 1223–12237.
- Griffiths et al. 1999. Modern Genetic Analysis. W. H. Freeman and Company.