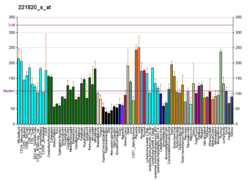
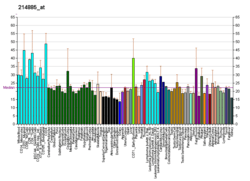
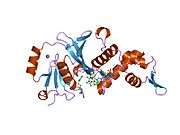
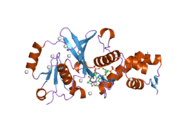KAT8
K(lysine) acetyltransferase 8 (KAT8) is an enzyme that in humans is encoded by the KAT8 gene.[5][6]
Function
The MYST family of histone acetyltransferases, which includes KAT8, was named for the founding members MOZ (MYST3; MIM 601408), yeast YBF2 and SAS2, and TIP60 (HTATIP; MIM 601409). All members of this family contain a MYST region of about 240 amino acids with a canonical acetyl-CoA-binding site and a C2HC-type zinc finger motif. Most MYST proteins also have a chromodomain involved in protein-protein interactions and targeting transcriptional regulators to chromatin.[6]
Interactions
KAT8 has been shown to interact with MORF4L1.[7]
gollark: Obviously the current george situation is quite troubling.
gollark: CSS makes it really easy and pleasant to do some things, and then nightmarishly horrible to do others.
gollark: Sort of? I mean, media queries. I don't think you can do that more generally.
gollark: Yes. However, there exists CSS grid.
gollark: Too late, I have an approximate mental model of it.
References
- GRCh38: Ensembl release 89: ENSG00000103510 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000030801 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Neal KC, Pannuti A, Smith ER, Lucchesi JC (Jan 2000). "A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1490 (1–2): 170–4. doi:10.1016/s0167-4781(99)00211-0. PMID 10786633.
- "Entrez Gene: MYST1 MYST histone acetyltransferase 1".
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM (Dec 2002). "MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation". The Journal of Biological Chemistry. 277 (52): 50860–6. doi:10.1074/jbc.M203839200. PMID 12397079.
Further reading
- Rea S, Xouri G, Akhtar A (Aug 2007). "Males absent on the first (MOF): from flies to humans". Oncogene. 26 (37): 5385–94. doi:10.1038/sj.onc.1210607. PMID 17694080.
- Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Kitabayashi I, Aikawa Y, Nguyen LA, Yokoyama A, Ohki M (Dec 2001). "Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein". The EMBO Journal. 20 (24): 7184–96. doi:10.1093/emboj/20.24.7184. PMC 125775. PMID 11742995.
- Pelletier N, Champagne N, Stifani S, Yang XJ (Apr 2002). "MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2". Oncogene. 21 (17): 2729–40. doi:10.1038/sj.onc.1205367. PMID 11965546.
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM (Dec 2002). "MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation". The Journal of Biological Chemistry. 277 (52): 50860–6. doi:10.1074/jbc.M203839200. PMID 12397079.
- Wan D, Gong Y, Qin W, Zhang P, Li J, Wei L, Zhou X, Li H, Qiu X, Zhong F, He L, Yu J, Yao G, Jiang H, Qian L, Yu Y, Shu H, Chen X, Xu H, Guo M, Pan Z, Chen Y, Ge C, Yang S, Gu J (Nov 2004). "Large-scale cDNA transfection screening for genes related to cancer development and progression". Proceedings of the National Academy of Sciences of the United States of America. 101 (44): 15724–9. doi:10.1073/pnas.0404089101. PMC 524842. PMID 15498874.
- Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK (Jun 2005). "Involvement of human MOF in ATM function". Molecular and Cellular Biology. 25 (12): 5292–305. doi:10.1128/MCB.25.12.5292-5305.2005. PMC 1140595. PMID 15923642.
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG (Jun 2005). "Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF". Cell. 121 (6): 873–85. doi:10.1016/j.cell.2005.04.031. PMID 15960975.
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A (Aug 2005). "hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells". Molecular and Cellular Biology. 25 (15): 6798–810. doi:10.1128/MCB.25.15.6798-6810.2005. PMC 1190338. PMID 16024812.
- Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M (Sep 2005). "Acetylation of HIV-1 integrase by p300 regulates viral integration". The EMBO Journal. 24 (17): 3070–81. doi:10.1038/sj.emboj.7600770. PMC 1201351. PMID 16096645.
- Smith ER, Cayrou C, Huang R, Lane WS, Côté J, Lucchesi JC (Nov 2005). "A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16". Molecular and Cellular Biology. 25 (21): 9175–88. doi:10.1128/MCB.25.21.9175-9188.2005. PMC 1265810. PMID 16227571.
- Topper M, Luo Y, Zhadina M, Mohammed K, Smith L, Muesing MA (Mar 2007). "Posttranslational acetylation of the human immunodeficiency virus type 1 integrase carboxyl-terminal domain is dispensable for viral replication". Journal of Virology. 81 (6): 3012–7. doi:10.1128/JVI.02257-06. PMC 1865993. PMID 17182677.
External links
- Overview of all the structural information available in the PDB for UniProt: Q9H7Z6 (Human Histone acetyltransferase KAT8) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: Q9D1P2 (Mouse Histone acetyltransferase KAT8) at the PDBe-KB.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.