MAGIChip
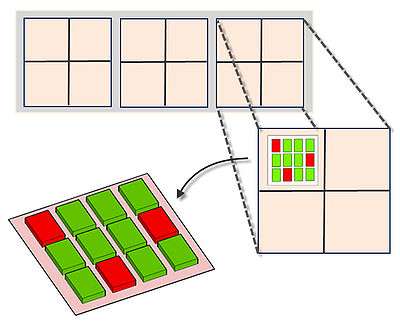
MAGIChips, also known as "microarrays of gel-immobilized compounds on a chip" or "three-dimensional DNA microarrays", are devices for molecular hybridization produced by immobilizing oligonucleotides, DNA, enzymes, antibodies, and other compounds on a photopolymerized micromatrix of polyacrylamide gel pads of 100x100x20µm or smaller size. This technology is used for analysis of nucleic acid hybridization, specific binding of DNA, and low-molecular weight compounds with proteins, and protein-protein interactions.
The gel pads increase the surface for hybridization to 50 times, compared to typical microarrays which are printed on flat surface of a glass slide that is usually treated by chemical compounds on which the probes adhere. A probe density of more than 1012 molecules per gel pad can be achieved due to 3D nature of the gel pads. The array is based on a glass surface that has small polyacrylamide gel units affixed to it. Each gel unit functions as an individual reaction cell as it is surrounded by a hydrophobic glass surface that prevents mixing of the solution in the gel units. This lays a foundation for performing ligation, single base extension, PCR amplification of DNA, on-chip MALDI-TOF mass spectrometry and other reactions.
Historical background
MAGIChip technology was developed as a result of collaboration between Dr. David Stahl at University of Washington and Dr Andrei Mirzabekov, formerly of Argonne National laboratory. Andrei Mirzabekov initiated the development of the DNA sequencing by hybridization with oligonucleotides: a novel method in 1988. This method was a foundation for the biotechnology that uses biological microchips to identify DNA structures rapidly, which is of great importance in the fight against a variety of diseases.
A joint research project was announced in 1998 among Motorola Inc, Packard Instrument Company and the U.S. Department of Energy's Argonne National Laboratory. In 1999, the researchers at Argonne National Lab pushed the development of microarray-type biochip technology they co-designed with the Engelhardt Institute to ward off a worldwide outbreak of tuberculosis.
Motorola developed manufacturing processes to mass-produce biochips, and Packard developed and manufactured the analytical instruments to process and analyze the biochips. Argonne's contribution, in conjunction with Engelhardt Institute of Molecular Biology (EIMB), was intellectual property in the form of 19 inventions related to biological microchips.
But this collaboration between EIMB in Moscow and Argonne National Laboratory at Illinois and two other US-based commercial partners collapsed as result of argument on contractual arrangement between the parties in 2001. As a result of this dispute, Dr Andrei Mirzabekov resigned as a director of Argonne's Biochip Technology Centre.
Method
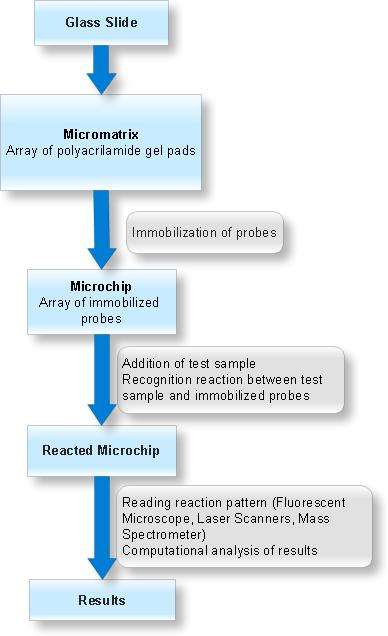
Arrays of gel elements (pads) are created on the glass surface (micromatrix) which is followed by application and chemical immobilization of different compounds (probes) onto these gel pads. Test sample is then added to this micromatrix containing immobilized probes in gel pads and molecular recognition reactions are allowed to take place under specified conditions. The test sample is fluorescent labelled to monitor the molecular interactions. The analysis of molecular interaction patterns is done by using specialized software.
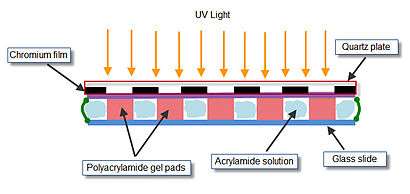
The array of gel elements on a glass slide is prepared by ‘’’photopolymerization‘’’. The acrylamide solution to be polymerized is applied to the polymerization chamber. Polymerization chamber consists of a quartz mask, two Teflon spacers, and a microscopic glass slide, clamped together by two metal clamps. The inner side of quartz mask has ultraviolet (UV)-transparent windows arranged in a specified spatial manner in a non-transparent chromium film. Assembled chamber containing the acrylamide gel is exposed to UV light to allow polymerization in only those positions of the chamber that are situated directly under the transparent windows.
Oligonucleotides or DNA fragments need to be activated to contain chemically reactive groups to facilitate coupling with the activated gel elements. Probe activation depends on the chemistry of activation of the polyacrylamide gels. Thus to immobilize in the aldehyde-containing gel the probe should have reactive amino group and if the gels are activated by introduction of amino groups, the probes should contain free aldehyde group. Probes are usually prepared by introduction of chemically active groups in terminal position of the oligonucleotides during their synthesis.
Probes for immobilization are transferred into gel elements of micromatrix by using dispensing robots. The fibre-optic pin of the robots has a hydrophobic side surface and a hydrophilic tip, and operates at a dew temperature to prevent evaporation of the sample during transfer. The activated probes are chemically immobilized by coupling oligonucleotides bearing amino or aldehyde groups with gel supports containing aldehyde or amino groups respectively.
The target molecules are labelled with fluorescent dyes. The fluorescent detection enables monitoring the process in real time with high spatial resolution. The criteria for labelling procedure includes –
- It should be simple, fast and inexpensive
- It should be applicable to both RNA and DNA targets
- It should be compatible with fragmentation required to decrease secondary structure formation
- It should allow incorporation of one label into one fragment to ensure proper quantification of the hybridization intensity
- It should allow coupling of multiple dyes
On-chip amplification reactions
On-chip amplification of the hybridization reaction serves as a very useful tool when the DNA or protein under study are present in relatively small proportion in the molecular population applied to the chip, e.g., when one is dealing with a single copy gene or mRNA of low abundance.
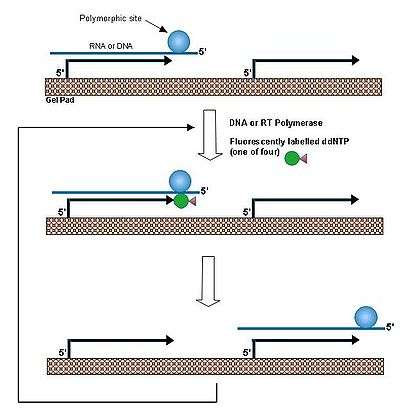
In a single base extension method,[1] a primer is hybridized to DNA and extended by a dideoxyribonucleoside triphosphate that matches the nucleotide at a polymorphic site. By performing this reaction at a temperature above the melting temperature of the duplex between the DNA and immobilized probe allows rapid association/dissociation of the target DNA. Thus the same DNA molecule reacts with many individual primers, leading to amplification of the primers in each individual gel pads. This procedure was applied to the identification of beta-globin gene mutation in the patients of beta thalassemia patients and to detection of anthrax toxin gene.
The chips also provides a good platform for performing PCR directly on the chip (in individual gel pads) as it is easy to isolate each gel pad from its neighbour unlike typical microarray chips which face serious problems in doing the same task
For the analysis of hybridization results obtained with fluorescently labelled target molecules fluorescence microscopes are employed. The instrument is equipped with controlled-temperature sample table to vary the temperature in the chip-containing reaction chamber during the course of the experiment. A cooled charge-coupled device (CCD) camera is used to record the light signals from the chip, which are then sent to the computer program for quantitative evaluation of the hybridization signals over the entire chip. Data generated by these experiments is stored in a database and analyzed by software that help provide evaluation, in silico experimentation, and hardware and software quality control.
Types
oligonucleotide chips
Customized oligonucleotide biochips are designed to interrogate test samples of known nucleotide sequences. For example, known genes in cases when one is interested in their expression levels under certain conditions, genes that are known to contain point mutations, or to be polymorphic in a given population. The success of the microarray depends on the proper choice of probes in these cases.
A set of potential hybridization probes are created for each DNA sequence that form perfect duplexes with that sequence. The potential probes that may create ambiguities in the interpretation of the hybridization pattern are excluded on the basis of AT vs GC content, and the propensity to form hairpins and other types of stable secondary structures that may drastically affect the intensity of hybridization.
One of the cases of successful applications of customized oligonucleotide chips include detection of beta-thalassemia mutation in patients. For the diagnostics of beta-thalassemia mutations, a simple chip was designed that contained six probes corresponding to different beta-thalassemia genotypes and hybridized with PCR-amplified DNA from healthy humans and patients.[2] The hybridization results showed the expected significant differences in signal intensity between matched and mismatched duplexes, thus allowing reliable identification of both homozygous and heterozygous mutations.
rRNA chips
These chips haves been developed for ribosomal RNA (rRNA) targets, commonly used for detecting bacteria. rRNA are very abundant in the cell comprising about 80% of the RNA content of the typical eukaryotic cell.[3] The rRNA is pre-amplified by bacteria and there are present in several thyousand copies per cell, making a good target for microassays. Single nucleotide polymorphisms present in the bacterial rRNA sequence are used to differentiate bacteria at the genus, species and strain level. This is a unique feature of this microchip that does not require PCR based amplification. The process for detecting bacterial is relatively simple. The bacteria are cultured, washed and pelleted. Lysosome is used to lyse the pellets - to destroy the cell walls and release the nucleic acid. Lysed bacteria are passed through a colourmn preparation where nucleic acid from the cell is immobilized and other debris is washed out. All the processes after lysis - isolation, purification, fragmentation and labelling of target rRNA's - are stable chemical reactions Fragments <500 bp easily hybridize to the gel matrix. The total number of eluted off the chip is determined by UV spectrophotometer. The process from sample preparation to identification of organisms based on automated algorithms take place in 2 hours.
cDNA
cDNAs obtained from reverse transcription of mRNA population extracted from the cells in varying physiological and experimental conditions are used as immobilized probes. These arrays are widely used to study gene expression. The potential obstacle in using cDNAs is due to the difficulty of injecting and evenly distributing long molecules into the gel pads. This problem is resolved by developing polyacrylamide gels that contain larger average pore size. Another way to approach this problem is to randomly fragment the cDNA into relatively small pieces before immobilization
Proteins
Protein chips can be prepared that contain different proteins immobilized as probes in a way that preserves their biological activity.[4] A large pore gel is used to prevent the diffusion of protein into the gel. There are two ways to immobilize proteins to the gel pads. The first is based on activation of the gel with glutyraldehyde. In the second procedure the gel is activated by partial substitution of amino groups with hydrazide groups. The reaction between hydrazide and aldehyde groups efficiently cross-links the protein to the cell. Protein microchips show the high specificity in molecular recognition reactions as seen in solution. Interaction between antigen and their specific antibodies can be studied on-chip in variety of experimental conditions. Either the antigen or antibody can be immobilized and monitored by both direct and indirect methods. In direct method, one uses target molecules labelled with fluorescent dye and in the indirect method the reaction is detected using the labelled molecule that specifically recognizes the target. These chips can be used to study enzymatic activity of immobilized enzymes by coating the chip with solution containing specific substrates. The reaction is monitored by detecting the formation of coloured or fluorescent precipitates
Other applications
- They can be used to study singlenucleotide polymorphisms (SNPs) in its method. Because bacterial DNA is highly conserved over time, SNPs are useful to identify bacterial on the chip, and since SNPs are the most abundant variations in the human genome, they have become the primary markers for genetic studies for mapping and identifying susceptible genes for complex diseases.[5]
- They can be used to detect virulence factors which are toxins and proteins that invade the organism. These toxins tend to have small number of copy transcripts and produced under very specific conditions found in the host. Here identification strategy focus on the single copy DNA sequence where MAGIChips are very effective.
- The protein biochips makes it very exciting as proteins are contained within a single cell and they can all be analyzed in one array platform. Protein biochips can be used to identify protein biomarkers for diagnosing diseases or a particular stage of the disease. They can also help to delineate relationship between protein structure and function of the protein and identify the function of a protein or different proteins throughout the same or different cell types. Although the MAGIChips need some modifications, the applications and techniques are quite standard.[6]
- The chips can be used as a diagnostic tool in clinics by virtue of its rapid detection time, high-throughput, result confidence, hierarchical identification and quantification. With them it is possible to achieve time required for sample collection to reporting of results in clinical settings within 2 hours. The rapid turnaround time is an attractive attribute of point-of-care testing while patient awaits the results.
- High-throughput nature of these devices allow thousands of microbial probes for species specific and even strain-specific identification at the same time on a single chip, reducing the amount of sample needed to conduct multiple tests. Potential threats posed by use of bacteria, viruses and fungi as biological weapons against humans, agriculture and environment warrants development of technology for accurate and sensitive detection within a very short time. MAGIChip prospective technology which has been used for discrimination of important viruses. Fungal probes have been introduced into rRNA chips for agricultural research in genetics, reproduction, diseases and even crop protection. Thousands of gene can be targeted simultaneously to look for genetic diversity or microbial infestation by nature or by intentional release.
See also
- Biochip
- Protein Microarrays
- Antibody microarray
- Cellular Microarray
- Chemical Compound Microarray
- DNA Microarray
- MicroArray and Gene Expression
- Lab-on-a-Chip
References
- (1998)The runners-up. Science 282, 2156–2157.
- Yershov, G., Barsky, V., Belgovskiy, A., Kirillov, E., Kreindlin, E., Ivanov, I., Parinov, S., Guschin, D., Drobyshev, A., Dubiley, S., and Mirzabekov, A. (1996) DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93, 4913–4918.
- Zlatanova, J., and Mirzabekov, A., “Gel-Immobilized Microarrays of Nucleic Acids and Proteins. Production and Application for Macromolecular Research,” Methods Mol. Biol. 170, 17−38 (2001).
- Guschin, D., Yershov, G., Zaslavsky, A., Gemmell, A., Shick, V., Proudnikov, D., Arenkov, P., and Mirzabekov, A. (1997) Manual manufacturing of oligonucleotide, DNA and protein microchips. Anal. Biochem. 250, 203–211.
- Weiner, M. P., and Hudson, T. J., “Introduction to SNPs: Discovery of Markers for Disease,” Biotechniques 32(S), 4−13 (2002).
- Biotechnology Industry Organization (Bio), The Technologies and Their Applications, available at http://www.bio.org/er/applications.asp.