Lipid-gated ion channels
Lipid-gated ion channels are a class of ion channels whose conductance of ions through the membrane depends directly on lipids. Classically the lipids are membrane resident anionic signaling lipids that bind to the transmembrane domain on the inner leaflet of the plasma membrane with properties of a classic ligand. Other classes of lipid-gated channels include the mechanosensitive ion channels that respond to lipid tension, thickness, and hydrophobic mismatch. A lipid ligand differs from a lipid cofactor in that a ligand derives its function by dissociating from the channel while a cofactor typically derives its function by remaining bound.[1]
| Lipid-gated ion-channel Kir2.2 | |
|---|---|
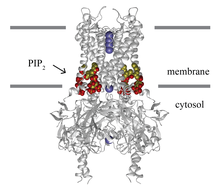 Tetrameric Kir2.2 (grey trace) bound to four PIP2 molecules (carbon:yellow; oxygen:red). Potassium ions (purple) are shown in the open conduction pathway. Grey rectangles indicate the membrane border. | |
| Identifiers | |
| Symbol | Kir2.2 |
| OPM protein | 3SPG |
PIP2-gated channels
Phosphatidylinositol 4,5-bisphosphate (PIP2) was the first and remains the best studied lipid to gate ion channels. PIP2 is a cell membrane lipid, and its role in gating ion channels represents a novel role for the molecule,[1][2]
Kir channels: PIP2 binds to and directly activates inwardly rectifying potassium channels (Kir).[3] The lipid bound in a well-defined ligand binding site in the transmembrane domain and cause the helices to splay opening the channel. All members of the Kir super-family of potassium channels are thought to be directly gated by PIP.[1]
Kv7 channels: PIP2 binds to and directly activates Kv7.1.[4]. In the same study PIP2 was shown to function as a ligand. When the channel was reconstituted into lipid vesicles with PIP2 the channel opened, when PIP2 was omitted the channel was closed.[4]
TRP channels: TRP channels were perhaps the first class of channels recognized as lipid-gated[5]. PIP2 regulates the conductance of most TRP channels either positively or negatively. For TRPV5, binding of PIP2 to a site in the transmembrane domain caused a conformational change that appeared to open the conduction pathway,[6] suggesting the channel is classically lipid-gated. A PIP2 compatible site was found in TRPV1 but whether the lipid alone can gate the channels has not been shown.[2] Other TRP channels that directly bind PIP2 are TRPM8 and TRPML.[7][8] Direct binding does not exclude PIP2 from affecting the channel by indirect mechanisms.
PA-gated channels
Phosphatidic acid (PA) recently emerged as an activator of ion channels.[9]
K2p: PA directly activates TREK-1 potassium channels through a putative site in the transmembrane domain. The affinity of PA for TREK-1 is relatively weak but the enzyme PLD2 produces high local concentration of PA to activate the channel.[10][11]
nAChR: PA also activates the nAChR in artificial membranes. Initially, the high concentration of PA required to activate nAChR[12] suggested a related anionic lipid might activate the channel, however, the finding of local high concentration of PA activating TREK-1 may suggest otherwise.
Kv: PA binding can also influence the midpoint of voltage activation (Vmid) for voltage-activated potassium channels.[13] Depletion of PA shifted the Vmid -40 mV near resting membrane potential which could open the channel absent a change in voltage suggesting these channels may also be lipid-gated. PA lipids were proposed to non-specifically gated a homologous channel from bacteria KvAP,[14] but those experiments did not rule out the anionic lipid phosphatidylglycerol from contributing specifically to gating.
PG-gated channels
Phosphatidylglycerol(PG) is an anionic lipid that activates many channels including most of the PA activated channels. The physiological signaling pathway is not well studied, but PLD can produce PG in the presence of glycerol[15] suggesting the same mechanism that is thought to generate local PA gradients could be generating high local PG gradients as well.
Mechanosensitive channels
A specialized set of mechanosensitive ion channels is gated by lipid deformation in the membrane in response to mechanical force. A theory involving the lipid membrane, called "force from lipid", is thought to directly open ion channels.[16] These channels include the bacterial channels MscL and MscS which open in response to lytic pressure. Many mechanosensitive channels require anionic lipids for activity[17].
Channels can also respond to membrane thickness. An amphipathic helix that runs along the inner membrane of TREK-1 channels is thought to sense changes in membrane thickness and gate the channel.[18]
Activation by localized lipid production
When an enzyme forms a complex with a channel it is thought to produce ligand near the channel in concentrations that are higher than the ligand in bulk membranes. [10] Theoretical estimates suggest initial concentration of a signaling lipid produced near an ion channel are likely millimolar;[9] however, due to theoretical calculations of lipids diffusion in a membrane, the ligand was thought to diffuse away much to fast to activate a channel.[19] However, Comoglio and colleagues showed experimentally that the enzyme phospholipase D2 bound directly to TREK-1 and produced the PA necessary to activate the channel.[10] The conclusion of Comoglio et al was experimentally confirmed when it was shown that the dissociation constant of PA for TREK-1 is 10 micro molar,[11] a Kd much weaker than the bulk concentration in the membrane. Combined these data show that PA must be local in concentration near 100 micro molar or more, suggesting the diffusion of the lipid is somehow restricted in the membrane.
Activation by membrane protein translocation
In theory, ion channels can be activated by their diffusion or trafficking to high concentrations of a signaling lipid.[9] The mechanism is similar to producing local high concentrations of a signaling lipid, but instead of changing the concentration of the lipid in the membrane near the channel, the channel moves to a region of the plasma membrane that already contains high concentrations of a signaling lipid. The change the channel experiences in lipid composition can be much faster and without any change in the total lipid concentration in the membrane.
Lipid competition
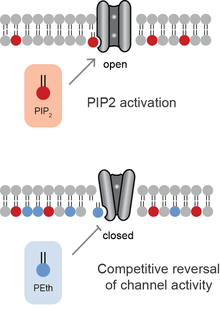
Anionic lipids compete for binding sites within ion channel. Similar to neurotransmitters, competition of an antagonist reverses the effect of an agonist. In most cases, the PA has the opposite effect of PIP2.[9] Hence when PA binds to a channel that is activated by PIP2, PA inhibits the effect of PIP2. When PA activates the channel, PIP2 blocks the effect of PA inhibiting the channels.
Ethanol When ethanol is consumed, phospholipase D incorporates the ethanol into phospholipids generating the unnatural and long lived lipid phosphatidylethanol(PEth) in a process called transphoshatidylation. The PEth competes with PA and the competition antagonizes TREK-1 channels. The competition of PEth on potassium channel is thought to contribute to the anesthetic effect of ethanol and perhaps hangover.[20]
References
- Hansen SB (May 2015). "Lipid agonism: The PIP2 paradigm of ligand-gated ion channels". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1851 (5): 620–8. doi:10.1016/j.bbalip.2015.01.011. PMC 4540326. PMID 25633344.
- Gao Y, Cao E, Julius D, Cheng Y (June 2016). "TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action". Nature. 534 (7607): 347–51. Bibcode:2016Natur.534..347G. doi:10.1038/nature17964. PMC 4911334. PMID 27281200.
- Hansen SB, Tao X, MacKinnon R (August 2011). "Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2". Nature. 477 (7365): 495–8. Bibcode:2011Natur.477..495H. doi:10.1038/nature10370. PMC 3324908. PMID 21874019.
- Sun J, MacKinnon R (January 2020). "Structural Basis of Human KCNQ1 Modulation and Gating". Cell. 180 (2): 340–347.e9. doi:10.1016/j.cell.2019.12.003. PMC 7083075. PMID 31883792.
- Benham, CD; Davis, JB; Randall, AD (June 2002). "Vanilloid and TRP channels: a family of lipid-gated cation channels". Neuropharmacology. 42 (7): 873–88. doi:10.1016/s0028-3908(02)00047-3. PMID 12069898.
- Hughes TE, Pumroy RA, Yazici AT, Kasimova MA, Fluck EC, Huynh KW, et al. (October 2018). "Structural insights on TRPV5 gating by endogenous modulators". Nature Communications. 9 (1): 4198. Bibcode:2018NatCo...9.4198H. doi:10.1038/s41467-018-06753-6. PMC 6179994. PMID 30305626.
- Fine M, Schmiege P, Li X (October 2018). "2-mediated human TRPML1 regulation". Nature Communications. 9 (1): 4192. doi:10.1038/s41467-018-06493-7. PMC 6180102. PMID 30305615.
- Yin Y, Le SC, Hsu AL, Borgnia MJ, Yang H, Lee SY (March 2019). "Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel". Science. 363 (6430): eaav9334. doi:10.1126/science.aav9334. PMC 6478609. PMID 30733385.
- Robinson CV, Rohacs T, Hansen SB (September 2019). "Tools for Understanding Nanoscale Lipid Regulation of Ion Channels". Trends in Biochemical Sciences. 44 (9): 795–806. doi:10.1016/j.tibs.2019.04.001. PMC 6729126. PMID 31060927.
- Comoglio Y, Levitz J, Kienzler MA, Lesage F, Isacoff EY, Sandoz G (September 2014). "Phospholipase D2 specifically regulates TREK potassium channels via direct interaction and local production of phosphatidic acid". Proceedings of the National Academy of Sciences of the United States of America. 111 (37): 13547–52. Bibcode:2014PNAS..11113547C. doi:10.1073/pnas.1407160111. PMC 4169921. PMID 25197053.
- Cabanos C, Wang M, Han X, Hansen SB (August 2017). "2 Antagonism of TREK-1 Channels". Cell Reports. 20 (6): 1287–1294. doi:10.1016/j.celrep.2017.07.034. PMC 5586213. PMID 28793254.
- Hamouda AK, Sanghvi M, Sauls D, Machu TK, Blanton MP (April 2006). "Assessing the lipid requirements of the Torpedo californica nicotinic acetylcholine receptor". Biochemistry. 45 (13): 4327–37. doi:10.1021/bi052281z. PMC 2527474. PMID 16566607.
- Hite RK, Butterwick JA, MacKinnon R (October 2014). "Phosphatidic acid modulation of Kv channel voltage sensor function". eLife. 3. doi:10.7554/eLife.04366. PMC 4212207. PMID 25285449.
- Zheng H, Liu W, Anderson LY, Jiang QX (22 March 2011). "Lipid-dependent gating of a voltage-gated potassium channel". Nature Communications. 2 (1): 250. Bibcode:2011NatCo...2..250Z. doi:10.1038/ncomms1254. PMC 3072105. PMID 21427721.
- Yang SF, Freer S, Benson AA (February 1967). "Transphosphatidylation by phospholipase D". The Journal of Biological Chemistry. 242 (3): 477–84. PMID 6022844.
- Teng J, Loukin S, Anishkin A, Kung C (January 2015). "The force-from-lipid (FFL) principle of mechanosensitivity, at large and in elements". Pflugers Archiv. 467 (1): 27–37. doi:10.1007/s00424-014-1530-2. PMC 4254906. PMID 24888690.
- Powl AM, East JM, Lee AG (April 2008). "Anionic phospholipids affect the rate and extent of flux through the mechanosensitive channel of large conductance MscL". Biochemistry. 47 (14): 4317–28. doi:10.1021/bi702409t. PMC 2566799. PMID 18341289.
- Nayebosadri A, Petersen EN, Cabanos C, Hansen SB (2018). "A Membrane Thickness Sensor in TREK-1 Channels Transduces Mechanical Force". doi:10.2139/ssrn.3155650. Cite journal requires
|journal=(help) - Hilgemann DW (October 2007). "Local PIP(2) signals: when, where, and how?". Pflugers Archiv. 455 (1): 55–67. doi:10.1007/s00424-007-0280-9. PMID 17534652.
- Chung HW, Petersen EN, Cabanos C, Murphy KR, Pavel MA, Hansen AS, et al. (January 2019). "A Molecular Target for an Alcohol Chain-Length Cutoff". Journal of Molecular Biology. 431 (2): 196–209. doi:10.1016/j.jmb.2018.11.028. PMC 6360937. PMID 30529033.