Kasugamycin
Kasugamycin (Ksg) is an aminoglycoside antibiotic that was originally isolated in 1965, from Streptomyces kasugaensis, a Streptomyces strain found near the Kasuga shrine in Nara, Japan. Kasugamycin was discovered by Hamao Umezawa, who also discovered kanamycin and bleomycin, as a drug that prevent growth of a fungus causing rice blast disease. It was later found to inhibit bacterial growth also. It exists as a white, crystalline substance with the chemical formula C14H28ClN3O10 (kasugamycin hydrochloride). It is also known as kasumin.[1]
 | |
| Names | |
|---|---|
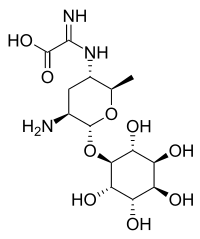
| IUPAC name
2-amino-2-[(2R,3S,5S,6R)-5-amino-2-methyl-6-[(2R,3S,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxyoxan-3-yl]iminoacetic acid | |
| Other names
Kasumin; 3-O-[2-Amino-4-[(carboxyiminomethyl)amino]-2,3,4,6-tetradeoxy-D-arabino-hexopyranosyl]-D-chiro-inositol | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
| ECHA InfoCard | 100.116.563 |
| KEGG | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C14H25N3O9 | |
| Molar mass | 379.366 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Mechanism of action
Like many of the known natural antibiotics, kasugamycin inhibits proliferation of bacteria by tampering with their ability to make new proteins, the ribosome being the major target. Kasugamycin inhibits protein synthesis at the step of translation initiation. Kasugamycin inhibition is thought to occur by direct competition with initiator transfer RNA. Recent experiments suggest that kasugamycin indirectly induces dissociation of P-site-bound fMet-tRNAfMet from 30S subunits through perturbation of the mRNA, thereby interfering with translation initiation.
Kasugamycin specifically inhibits translation initiation of canonical but not of leaderless mRNA. For initiation on leaderless mRNA, the overlap between mRNA and kasugamycin is reduced and the binding of tRNA is further stabilized by the presence of the 50S subunit, minimizing Ksg efficacy. Kasugamycin also induces the formation of unusual 61S ribosomes in vivo, which are proficient in selectively translating leaderless mRNA. 61S particles are stable and are devoid of more than six proteins of the small subunit, including the functionally important proteins S1 and S12.
Structural basis for kasugamycin action
The structure of the kasugamycin-70S ribosome complex from Escherichia coli has been determined by X-ray crystallography at 3.5-A resolution. The drug binds within the messenger RNA channel of the 30S subunit between the universally conserved G926 and A794 nucleotides in 16S ribosomal RNA, which are sites of kasugamycin resistance. The kasugamycin binding sites are present on top of helix 44 (h44), spanning the region between h24 and h28, which contacts the conserved nucleotides A794 and G926. Neither binding position overlaps with P-site tRNA. Instead, kasugamycin mimics the codon nucleotides at the P and E sites by binding within the path of the mRNA, thus perturbing the mRNA-tRNA codon-anticodon interaction.
Resistance
Low level resistance to kasugamycin is acquired by mutations in the 16S rRNA methyltransferase KsgA which methylates the nucleotides A1518 and A1519 in 16S rRNA. Spontaneous ksgA mutations conferring a modest level of resistance to kasugamycin occur at a high frequency of 10−6. Once cells acquire the ksgA mutations, they produce high-level kasugamycin resistance at an extraordinarily high frequency (100-fold greater frequency than that observed in the ksgA+ strain).
Surprisingly, kasugamycin resistance mutations do not inhibit binding of the drug to the ribosome. Present structural and biochemical results indicate that inhibition by kasugamycin and kasugamycin resistance are closely linked to the structure of the mRNA at the junction of the peptidyl-tRNA and exit-tRNA sites (P and E sites).
References
- Franz Müller, Peter Ackermann, Paul Margot (2012). "Fungicides, Agricultural, 2. Individual Fungicides". Ullmann's Encyclopedia of Industrial Chemistry. Weinheim: Wiley-VCH. doi:10.1002/14356007.o12_o06.CS1 maint: uses authors parameter (link)
Further reading
1. Okuyama, A., Machiyama, N., Kinoshita, T., and Tanaka, N. (1971). Inhibition by kasugamycin of initiation complex formation on 30S ribosomes. Biochem. Biophys. Res. Commun. 43, 196–199.
2. Schluenzen, F., Takemoto, C., Wilson, D.N., Kaminishi, T., Harms, J.M., Hanawa-Suetsugu, K., Szaflarski, W., Kawazoe, M., Shirouzu, M., Nierhaus, K.H., et al. (2006). The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat. Struct. Mol. Biol. 13, 871–878.
3. Schuwirth, B.S., Day, J.M., Hau, C.W., Janssen, G.R., Dahlberg, A.E., Cate, J.H., and Vila-Sanjurjo, A. (2006). Structural analysis of kasugamycin inhibition of translation. Nat. Struct. Mol. Biol. 13, 879–886.
4. Kaberdina A.C., Szaflarski W., Nierhaus K.H., and Moll I. (2009). An Unexpected Type of Ribosomes Induced by Kasugamycin: A Look into Ancestral Times of Protein Synthesis?. Mol. Cell. 33(2):141-2.
5. Ochi K., Kim J., Tanaka Y., Wang G., Masuda K., Nanamiya H., Okamoto S., Tokuyama S., Adachi Y. and Kawamura F. (2009). Inactivation of KsgA, a 16S rRNA Methyltransferase, Causes Vigorous Emergence of Mutants with High-Level Kasugamycin Resistance. Antimicrobial Agents and Chemotherapy, 53,1 (193-201).
6. Mankin A. (2006). Antibiotic blocks mRNA path on the ribosome. Nature Structural & Molecular Biology - 13, 858 – 860.