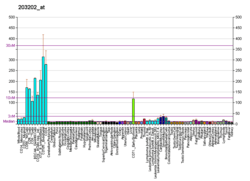
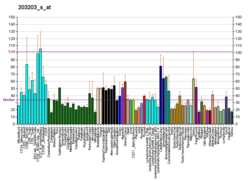
KRR1
KRR1 small subunit processome component homolog is a protein that in humans is encoded by the KRR1 gene.[5][6][7][8][9]
References
- GRCh38: Ensembl release 89: ENSG00000111615 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000063334 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Epplen C, Epplen JT (Feb 1994). "Expression of (cac)n/(gtg)n simple repetitive sequences in mRNA of human lymphocytes". Hum Genet. 93 (1): 35–41. doi:10.1007/BF00218910. PMID 7505766.
- Sasaki T, Toh-E A, Kikuchi Y (Oct 2000). "Yeast Krr1p Physically and Functionally Interacts with a Novel Essential Kri1p, and Both Proteins Are Required for 40S Ribosome Biogenesis in the Nucleolus". Mol Cell Biol. 20 (21): 7971–9. doi:10.1128/MCB.20.21.7971-7979.2000. PMC 86407. PMID 11027267.
- Chan HY, Brogna S, O'Kane CJ (May 2001). "Dribble, the Drosophila KRR1p Homologue, Is Involved in rRNA Processing". Mol Biol Cell. 12 (5): 1409–19. doi:10.1091/mbc.12.5.1409. PMC 34593. PMID 11359931.
- Gromadka R, Kaniak A, Slonimski PP, Rytka J (Aug 1996). "A novel cross-phylum family of proteins comprises a KRR1 (YCL059c) gene which is essential for viability of Saccharomyces cerevisiae cells". Gene. 171 (1): 27–32. doi:10.1016/0378-1119(96)00024-8. PMID 8675026.
- "Entrez Gene: KRR1 KRR1, small subunit (SSU) processome component, homolog (yeast)".
Further reading
- Muthumani K, Choo AY, Premkumar A, et al. (2006). "Human immunodeficiency virus type 1 (HIV-1) Vpr-regulated cell death: insights into mechanism". Cell Death Differ. 12 Suppl 1: 962–70. doi:10.1038/sj.cdd.4401583. PMID 15832179.
- Andersen JS, Lyon CE, Fox AH, et al. (2002). "Directed proteomic analysis of the human nucleolus". Curr. Biol. 12 (1): 1–11. doi:10.1016/S0960-9822(01)00650-9. PMID 11790298.
- Scherl A, Couté Y, Déon C, et al. (2003). "Functional Proteomic Analysis of Human Nucleolus". Mol. Biol. Cell. 13 (11): 4100–9. doi:10.1091/mbc.E02-05-0271. PMC 133617. PMID 12429849.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Micheau O, Tschopp J (2003). "Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes". Cell. 114 (2): 181–90. doi:10.1016/S0092-8674(03)00521-X. PMID 12887920.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Bernstein KA, Gallagher JE, Mitchell BM, et al. (2005). "The Small-Subunit Processome Is a Ribosome Assembly Intermediate". Eukaryotic Cell. 3 (6): 1619–26. doi:10.1128/EC.3.6.1619-1626.2004. PMC 539036. PMID 15590835.
- Andersen JS, Lam YW, Leung AK, et al. (2005). "Nucleolar proteome dynamics". Nature. 433 (7021): 77–83. doi:10.1038/nature03207. PMID 15635413.
- Rual JF, Venkatesan K, Hao T, et al. (2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Oh JH, Yang JO, Hahn Y, et al. (2006). "Transcriptome analysis of human gastric cancer". Mamm. Genome. 16 (12): 942–54. doi:10.1007/s00335-005-0075-2. PMID 16341674.
- Olsen JV, Blagoev B, Gnad F, et al. (2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.