k-medoids
The k-medoids or partitioning around medoids (PAM) algorithm is a clustering algorithm reminiscent of the k-means algorithm. Both the k-means and k-medoids algorithms are partitional (breaking the dataset up into groups) and both attempt to minimize the distance between points labeled to be in a cluster and a point designated as the center of that cluster. In contrast to the k-means algorithm, k-medoids chooses data points as centers (medoids or exemplars) and can be used with arbitrary distances, while in k-means the centre of a cluster is not necessarily one of the input data points (it is the average between the points in the cluster). The PAM method was proposed in 1987[1] for the work with norm and other distances.
k-medoid is a classical partitioning technique of clustering, which clusters the data set of n objects into k clusters, with the number k of clusters assumed known a priori (which implies that the programmer must specify k before the execution of the algorithm). The "goodness" of the given value of k can be assessed with methods such as the silhouette method.
It is more robust to noise and outliers as compared to k-means because it minimizes a sum of pairwise dissimilarities instead of a sum of squared Euclidean distances. (This statement is not correct if the dissimilarity is based on L2 norm; it is therefore misleading. Robustness comes not from not using means, but from using non L-2 metrics such as L-1 or cosine).
A medoid can be defined as the object of a cluster whose average dissimilarity to all the objects in the cluster is minimal, that is, it is a most centrally located point in the cluster.
Algorithms
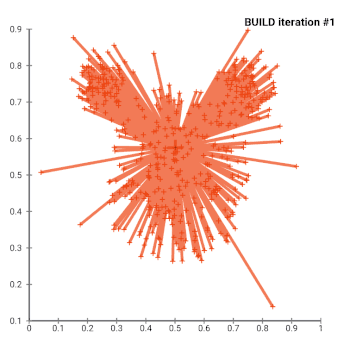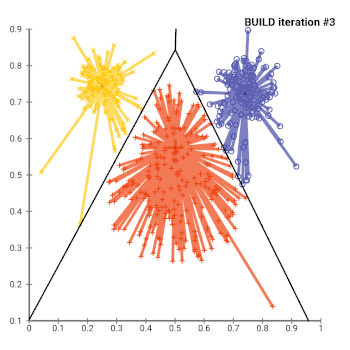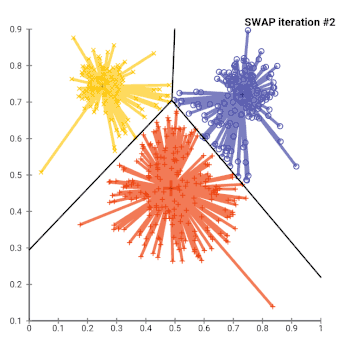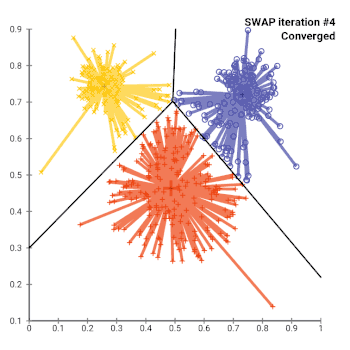
The most common realisation of k-medoid clustering is the partitioning around medoids (PAM) algorithm. PAM uses a greedy search which may not find the optimum solution, but it is faster than exhaustive search. It works as follows:
- Initialize: greedily select k of the n data points as the medoids to minimize the cost
- Associate each data point to the closest medoid.
- While the cost of the configuration decreases:
- For each medoid m, and for each non-medoid data point o:
- Consider the swap of m and o, and compute the cost change
- If the cost change is the current best, remember this m and o combination
- Perform the best swap of and , if it decreases the cost function. Otherwise, the algorithm terminates.
- For each medoid m, and for each non-medoid data point o:
The runtime complexity of the original PAM algorithm per iteration of (3) is , by only computing the change in cost. A naive implementation recomputing the entire cost function every time will be in . This runtime can be further reduced to , by splitting the cost change into three parts such that computations can be shared or avoided.[2]
Algorithms other than PAM have also been suggested in the literature, including the following Voronoi iteration method:[3][4][5]
- Select initial medoids randomly
- Iterate while the cost decreases:
- In each cluster, make the point that minimizes the sum of distances within the cluster the medoid
- Reassign each point to the cluster defined by the closest medoid determined in the previous step.
However, k-means-style Voronoi iteration finds worse results, as it does not allow reassigning points to other clusters while changing means, and thus only explores a smaller search space.[2][6]
The approximate algorithms CLARA and CLARANS trade optimality for runtime. CLARA applies PAM on multiple subsamples, keeping the best result. CLARANS works on the entire data set, but only explores a subset of the possible swaps of medoids and non-medoids using sampling.
Software
- ELKI includes several k-medoid variants, including a Voronoi-iteration k-medoids, the original PAM algorithm, Reynolds' improvements, and the O(n²) FastPAM algorithm, CLARA, CLARANS, FastCLARA and FastCLARANS.
- Julia contains a k-medoid implementation of the k-means style algorithm (faster, but much worse result quality) in the JuliaStats/Clustering.jl package.
- KNIME includes a k-medoid implementation supporting a variety of efficient matrix distance measures, as well as a number of native (and integrated third-party) k-means implementations
- R contains PAM in the "cluster" package, including some of the FastPAM improvements via the pamonce=5 option.
- RapidMiner has an operator named KMedoids, but it does not implement the KMedoids algorithm correctly. Instead, it is a k-means variant, that substitutes the mean with the closest data point (which is not the medoid).
- MATLAB implements PAM, CLARA, and two other algorithms to solve the k-medoid clustering problem.
References
- Kaufman, L. and Rousseeuw, P.J. (1987), Clustering by means of Medoids, in Statistical Data Analysis Based on the –Norm and Related Methods, edited by Y. Dodge, North-Holland, 405–416.
- Schubert, Erich; Rousseeuw, Peter J. (2019), Amato, Giuseppe; Gennaro, Claudio; Oria, Vincent; Radovanović, Miloš (eds.), "Faster k-Medoids Clustering: Improving the PAM, CLARA, and CLARANS Algorithms", Similarity Search and Applications, Springer International Publishing, 11807, pp. 171–187, arXiv:1810.05691, doi:10.1007/978-3-030-32047-8_16, ISBN 9783030320461
- Maranzana, F. E. (1963). "On the location of supply points to minimize transportation costs". IBM Systems Journal. 2 (2): 129–135. doi:10.1147/sj.22.0129.
- T. Hastie, R. Tibshirani, and J. Friedman. The Elements of Statistical Learning, Springer (2001), 468–469.
- Park, Hae-Sang; Jun, Chi-Hyuck (2009). "A simple and fast algorithm for K-medoids clustering". Expert Systems with Applications. 36 (2): 3336–3341. doi:10.1016/j.eswa.2008.01.039.
- Teitz, Michael B.; Bart, Polly (1968-10-01). "Heuristic Methods for Estimating the Generalized Vertex Median of a Weighted Graph". Operations Research. 16 (5): 955–961. doi:10.1287/opre.16.5.955. ISSN 0030-364X.