Intermembrane space
The intermembrane space (IMS) is the space occurring between or involving two or more membranes.[1] In cell biology, it is most commonly described as the region between the inner membrane and the outer membrane of a mitochondrion or a chloroplast. It also refers to the space between the inner and outer nuclear membranes of the nuclear envelope, but is often called the perinuclear space.[2] The IMS of mitochondria plays a crucial role in coordinating a variety of cellular activities, such as regulation of respiration and metabolic functions. Unlike the IMS of the mitochondria, the IMS of the chloroplast does not seem to have any obvious function.
.svg.png)
Intermembrane space of mitochondria
Mitochondria are surrounded by two membranes, consisting of outer and inner mitochondrial membranes. These two membranes allow the formation of two aqueous compartments, which are the intermembrane space (IMS) and the matrix.[3] Channel proteins called porins in the outer membrane allow free diffusion of ions and small proteins about 5000 daltons or less into the IMS. This makes the IMS chemically equivalent to the cytosol regarding the small molecules it contains. By contrast, specific transport proteins are required to transport ions and other small molecules across the inner mitochondrial membrane into the matrix due to its impermeability.[4] The IMS also contains many enzymes that use the ATP moving out of the matrix to phosphorylate other nucleotides and proteins that initiate apoptosis.[5]
Translocation
Most of proteins destined for the mitochondrial matrix are synthesized as precursors in the cytosol and are imported into the mitochondria by the translocase of the outer membrane (TOM) and the translocase of the inner membrane (TIM).[4][6] The IMS is involved in the mitochondrial protein translocation. The precursor proteins called small TIM chaperones which are hexameric complexes are located in the IMS and they bind hydrophobic precursor proteins and delivery the precursors to the TIM.[7]
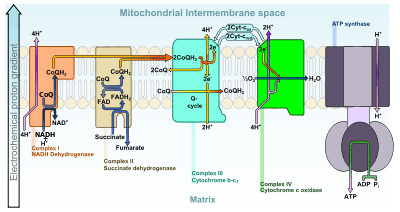
Oxidative phosphorylation
The pyruvate generated by glycolysis and the fatty acids produced by breakdown of fats enter the mitochondrial IMS through the porins in the outer mitochondrial membrane.[8] Then they are transported across the inner mitochondrial membrane into the matrix and converted into the acetyl CoA to enter the citric acid cycle.[8][9]
The respiratory chain in the inner mitochondrial membrane carries out oxidative phosphorylation. Three enzyme complexes are responsible for the electron transport: NADH-ubiquinone oxidoreductase complex (complex I), ubiquinone-cytochrome c oxidoreductase complex (complex III), and cytochrome c oxidase (complex IV).[10] The protons are pumped from the mitochondrial matrix to the IMS by these respiratory complexes. As a result, an electrochemical gradient is generated, which is combined by forces due to a H+ gradient (pH gradient) and a voltage gradient (membrane potential). The pH in the IMS is about 0.7 unit lower than the one in the matrix and the membrane potential of the IMS side becomes more positively charged than the matrix side. This electrochemical gradient from the IMS to the matrix is used to drive the synthesis of ATP in the mitochondria.[5]
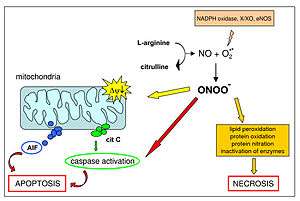
Apoptosis
Releasing of cytochrome c from the IMS to the cytosol activates procaspases and triggers a caspase cascade leading to apoptosis.[4]
Intermembrane space of chloroplasts
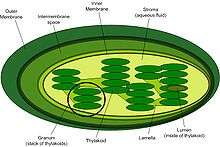
The intermembrane space (IMS) of the chloroplast is exceedingly small, from 10-20 nm thick. Unlike the IMS of the mitochondria, the IMS of the chloroplast does not seem to have any obvious function. The translocase of the outer membrane (TOC) and the translocase of the inner membrane (TIC) mainly assist the translocation of chloroplast precursor proteins[11] Chaperone involvement in the IMS has been proposed but still remains uncertain.The eukaryotic Hsp70, which is the heat shock protein of 70 kDa, typically localized in the cytoplasm is also found in the IMS of chloroplasts. The resulting hypothesis states that co-localization of Hsp70 is important for efficient translocation of protein precursors into and across the IMS of chloroplasts.[12]
Intermembrane space of nuclear envelopes
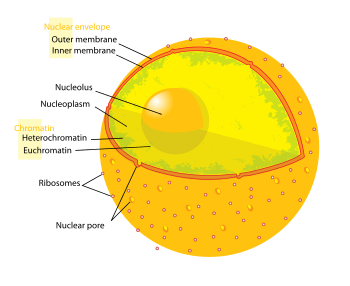
The nuclear envelope is composed of two lipid bilayer membranes that are penetrated by nuclear pores and separated by a small intermembrane space, which is often called the perinuclear space.[13] The perinuclear space is usually about 20-40 nm wide.[14] The perinuclear translocation of certain proteins and enzymes were studied and results showed that perinuclear space was important for genome integrity and gene regulation.[15]
References
- "Definition of INTERMEMBRANE". www.merriam-webster.com. Retrieved 2019-04-09.
- "Nuclear envelope", Wikipedia, 2019-03-24, retrieved 2019-04-01
- Cooper GM (2000). "Mitochondria". The Cell: A Molecular Approach (2nd ed.).
- Essential Cell Biology. Alberts, Bruce., Bray, Dennis., Hopkin, Karen., Johnson, Alexander D., Lewis, Julian. Garland Pub. 2014. ISBN 9780815345251. OCLC 881664767.CS1 maint: others (link)
- Manganas P, MacPherson L, Tokatlidis K (January 2017). "Oxidative protein biogenesis and redox regulation in the mitochondrial intermembrane space". Cell and Tissue Research. 367 (1): 43–57. doi:10.1007/s00441-016-2488-5. PMC 5203823. PMID 27632163.
- Pfanner N, Meijer M (February 1997). "The Tom and Tim machine". Current Biology. 7 (2): R100-3. doi:10.1016/S0960-9822(06)00048-0. PMID 9081657.
- Wiedemann N, Pfanner N (June 2017). "Mitochondrial Machineries for Protein Import and Assembly". Annual Review of Biochemistry. 86 (1): 685–714. doi:10.1146/annurev-biochem-060815-014352. PMID 28301740.
- Chaudhry R, Varacallo M (2019). "Biochemistry, Glycolysis". StatPearls. StatPearls Publishing. PMID 29493928. Retrieved 2019-04-09.
- "Structural Biochemistry/Krebs Cycle (Citric Acid cycle) - Wikibooks, open books for an open world". en.wikibooks.org. Retrieved 2019-04-09.
- Sousa JS, D'Imprima E, Vonck J (2018). "Mitochondrial Respiratory Chain Complexes". Subcellular Biochemistry. 87: 167–227. doi:10.1007/978-981-10-7757-9_7. ISBN 978-981-10-7756-2. PMID 29464561. Cite journal requires
|journal=(help) - Jarvis P, Soll J (December 2001). "Toc, Tic, and chloroplast protein import". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1541 (1–2): 64–79. doi:10.1016/S0167-4889(01)00147-1. PMID 11750663.
- Bionda T, Gross LE, Becker T, Papasotiriou DG, Leisegang MS, Karas M, Schleiff E (March 2016). "Eukaryotic Hsp70 chaperones in the intermembrane space of chloroplasts". Planta. 243 (3): 733–47. doi:10.1007/s00425-015-2440-z. PMID 26669598.
- Walter, Peter; Roberts, Keith; Raff, Martin; Lewis, Julian; Johnson, Alexander; Alberts, Bruce (2002). "The Transport of Molecules between the Nucleus and the Cytosol". Molecular Biology of the Cell. 4th Edition.
- "Perinuclear space - Biology-Online Dictionary | Biology-Online Dictionary". www.biology-online.org. Retrieved 2019-04-02.
- Shaiken TE, Opekun AR (May 2014). "Dissecting the cell to nucleus, perinucleus and cytosol". Scientific Reports. 4: 4923. Bibcode:2014NatSR...4E4923S. doi:10.1038/srep04923. PMC 4017230. PMID 24815916.