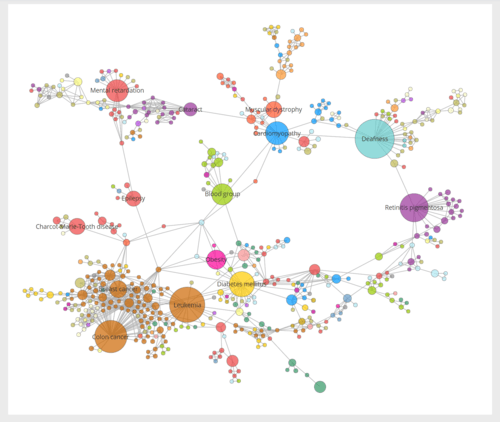
Human disease network
A human disease network is a network of human disorders and diseases with reference to their genetic origins or other features. More specifically, it is the map of human disease associations referring mostly to disease genes. For example, in a human disease network, two diseases are linked if they share at least one associated gene. A typical human disease network usually derives from bipartite networks which consist of both diseases and genes information. Additionally, some human disease networks use other features such as symptoms and proteins to associate diseases.
History
In 2007, Goh et al. constructed a disease-gene bipartite graph using information from OMIM database and termed human disease network.[2] In 2009, Barrenas et al. derived complex disease-gene network using GWAs (Genome Wide Association studies).[3] In the same year, Hidalgo et al. published a novel way of building human phenotypic disease networks in which diseases were connected according to their calculated distance.[4] In 2011, Cusick et al. summarized studies on genotype-phenotype associations in cellular context.[5] In 2014, Zhou, et al. built a symptom-based human disease network by mining biomedical literature database.[6]
Properties
A large-scale human disease network shows scale-free property. The degree distribution follows a power law suggesting that only a few diseases connect to a large number of diseases, whereas most diseases have few links to others. Such network also shows a clustering tendency by disease classes.[2][7]
In a symptom-based disease network, disease are also clustered according to their categories. Moreover, diseases sharing the same symptom are more likely to share the same genes and protein interactions.[6]
References
- http://nbviewer.jupyter.org/github/empet/Human-Disease-Network/blob/master/Human-Disease-Network.ipynb
- Goh, Kwang-Il, et al. "The human disease network." Proceedings of the National Academy of Sciences 104.21 (2007): 8685-8690.
- Barrenas, Fredrik, et al. "Network properties of complex human disease genes identified through genome-wide association studies." PLoS ONE 4.11 (2009): e8090.
- Hidalgo, César A., et al. "A dynamic network approach for the study of human phenotypes." PLoS Computational Biology 5.4 (2009): e1000353.
- Vidal, Marc, Michael E. Cusick, and Albert-Laszlo Barabasi. "Interactome networks and human disease." Cell 144.6 (2011): 986-998.
- Zhou, XueZhong, et al. "Human symptoms–disease network." Nature Communications 5 (2014).
- Kannan, Venkateshan et. al. "Conditional Disease Development extracted from Longitudinal Health Care Cohort Data using Layered Network Construction". Scientific Reports 6, 26170 (2016)