Helical wheel
A helical wheel is a type of plot or visual representation used to illustrate the properties of alpha helices in proteins.
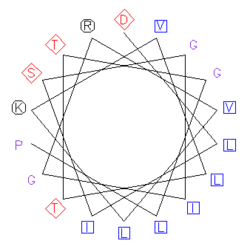
The sequence of amino acids that make up a helical region of the protein's secondary structure are plotted in a rotating manner where the angle of rotation between consecutive amino acids is 100°, so that the final representation looks down the helical axis.
Polarity and Characteristics
The plot reveals whether hydrophobic amino acids are concentrated on one side of the helix, usually with polar or hydrophilic amino acids on the other. This arrangement is common in alpha helices within globular proteins, where one face of the helix is oriented toward the hydrophobic core and one face is oriented toward the solvent-exposed surface. Specific patterns characteristic of protein folds and protein docking motifs are also revealed, as in the identification of leucine zipper dimerization regions and coiled coils. This projection diagram is often called and "Edmundson wheel" after its inventor.[1]
Drawing Helical Wheels
Helical wheels can be drawn by the HELIQUEST server and subsequently downloaded.[2] The helixvis package for the R programming language can also be used to draw customized helical wheels.[3]
References
- Schiffer M, Edmundson AB (1967). "Use of helical wheels to represent the structures of proteins and to identify segments with helical potential". Biophysical Journal. 7 (2): 121–35. Bibcode:1967BpJ.....7..121S. doi:10.1016/S0006-3495(67)86579-2. PMC 1368002. PMID 6048867.
- Gautier, R; Douguet, D; Antonny, B; Drin, G (2008). "HELIQUEST: a web server to screen sequences with specific alpha-helical properties". Bioinformatics. 24 (18): 2101–2102. doi:10.1093/bioinformatics/btn392. PMID 18662927.
- Wadhwa, R; Subramanian, V; Stevens-Truss, R (2018). "Visualizing alpha-helical peptides in R with helixvis". Journal of Open Source Software. 3 (31): 1008. Bibcode:2018JOSS....3.1008W. doi:10.21105/joss.01008.
Further reading
- Mount DM (2004). Bioinformatics: Sequence and Genome Analysis (2 ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. ISBN 0-87969-712-1.