HUH-tag
HUH endonucleases (HUH-tags) are sequence-specific single-stranded DNA (ssDNA) binding proteins originating from numerous species of bacteria and viruses.[1] Viral HUH endonucleases are naturally involved in initiating rolling circle replication while ones of bacterial origin initiate bacterial conjugation. In biotechnology, they can be used to create protein-DNA linkages,[2] akin to other methods such as SNAP-tag. In doing so, they create a 5' covalent bond between the ssDNA and the protein. HUH endonucleases can be fused with other proteins or used as protein tags.

Types of HUH endonucleases
HUH endonucleases are broadly split into two categories of enzymes: replication initiator proteins (Rep) or relaxase / mobilization proteins. They both contain small protein domains that recognize sequence-specific origins of replication or origin of transfer at which site they nick DNA. The nicking domain of Reps tend to be smaller, on the order of 10-20 kDa while nicking domains from relaxases are larger, roughly 20-40 kDa in size.[2]
Mode of action
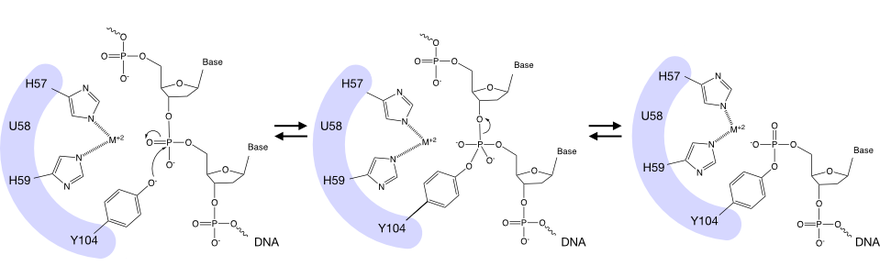
HUH endonucleases generally have two histidine (H) residues in the active site coordinating a metal cation (Mg2+ or Mn2+) that interacts with the phosphate backbone of DNA. This allows for a nucleophilic attack, most commonly, by an activated tyrosine of the scissile phosphate in the DNA backbone, generating a 5' covalent bond with the ssDNA. In contrast to other DNA-protein linkage approaches, this reaction occurs at ambient conditions and does not require any additional modifications. X-ray crystallography and NMR structures have provided insight into the sequence specificity of DNA binding.[3][4]
Applications
- MobA relaxase incorporated into the viral capsid of Adeno-associated virus to link a DNA-antibody conjugate to target the virus to specific cell types[5]
- PCV2 Rep protein fused to Cas9 to covalently link a DNA repair template to Cas9, resulting in increased homology-directed repair in human cells[6]
- Similar to the approach mentioned above, Agrobacterium VirD2 relaxase fused to Cas9 allowing for linking of a DNA repair template to increase homology-directed repair in plants[7]
- PCV2 Rep protein fused to Elastin-like particles (ELPs) linked to a Mucin-1 DNA aptamer to deliver drugs to cancer cells[8]
- TraI, MobA, and TrwC relaxases used in orthogonal assembly on DNA nanostructures[9]
- PCV2 Rep protein fused to luciferase linked to DNA aptamers that detected thrombin levels in a sample[10]
References
- Chandler, Michael; de la Cruz, Fernando; Dyda, Fred; Hickman, Alison B.; Moncalian, Gabriel; Ton-Hoang, Bao (2013-07-08). "Breaking and joining single-stranded DNA: the HUH endonuclease superfamily". Nature Reviews Microbiology. 11 (8): 525–538. doi:10.1038/nrmicro3067. ISSN 1740-1526. PMC 6493337. PMID 23832240.
- Lovendahl, Klaus N.; Hayward, Amanda N.; Gordon, Wendy R. (2017-05-24). "Sequence-Directed Covalent Protein–DNA Linkages in a Single Step Using HUH-Tags". Journal of the American Chemical Society. 139 (20): 7030–7035. doi:10.1021/jacs.7b02572. ISSN 0002-7863. PMC 5517037. PMID 28481515.
- Vega-Rocha, Susana; Byeon, In-Ja L.; Gronenborn, Bruno; Gronenborn, Angela M.; Campos-Olivas, Ramón (2007). "Solution Structure, Divalent Metal and DNA Binding of the Endonuclease Domain from the Replication Initiation Protein from Porcine Circovirus 2". Journal of Molecular Biology. 367 (2): 473–487. doi:10.1016/j.jmb.2007.01.002. ISSN 0022-2836. PMID 17275023.
- Everett, Blake A.; Litzau, Lauren A.; Tompkins, Kassidy; Shi, Ke; Nelson, Andrew; Aihara, Hideki; Evans Iii, Robert L.; Gordon, Wendy R. (2019-12-01). "Crystal structure of the Wheat dwarf virus Rep domain". Acta Crystallographica. Section F, Structural Biology Communications. 75 (Pt 12): 744–749. doi:10.1107/S2053230X19015796. ISSN 2053-230X. PMC 6891580. PMID 31797816.
- Zdechlik, Alina C.; He, Yungui; Aird, Eric J.; Gordon, Wendy R.; Schmidt, Daniel (2019-12-06). "Programmable Assembly of Adeno-Associated Virus–Antibody Composites for Receptor-Mediated Gene Delivery". Bioconjugate Chemistry. 31 (4): 1093–1106. doi:10.1021/acs.bioconjchem.9b00790. ISSN 1043-1802. PMID 31809024.
- Aird, Eric J.; Lovendahl, Klaus N.; Martin, Amber St; Harris, Reuben S.; Gordon, Wendy R. (2018-05-31). "Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template". Communications Biology. 1 (1): 54. doi:10.1038/s42003-018-0054-2. ISSN 2399-3642. PMC 6123678. PMID 30271937.
- Ali, Zahir; Shami, Ashwag; Sedeek, Khalid; Kamel, Radwa; Alhabsi, Abdulrahman; Tehseen, Muhammad; Hassan, Norhan; Butt, Haroon; Kababji, Ahad; Hamdan, Samir M.; Mahfouz, Magdy M. (2020-01-23). "Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice". Communications Biology. 3 (1): 44. doi:10.1038/s42003-020-0768-9. ISSN 2399-3642. PMC 6978410. PMID 31974493.
- Guo, Wei; Mashimo, Yasumasa; Kobatake, Eiry; Mie, Masayasu (2020-03-16). "Construction of DNA-displaying nanoparticles by enzymatic conjugation of DNA and elastin-like polypeptides using a replication initiation protein". Nanotechnology. 31 (25): 255102. doi:10.1088/1361-6528/ab8042. ISSN 0957-4484. PMID 32176872.
- Sagredo, Sandra; Pirzer, Tobias; Aghebat Rafat, Ali; Goetzfried, Marisa A.; Moncalian, Gabriel; Simmel, Friedrich C.; de la Cruz, Fernando (2016). "Orthogonal Protein Assembly on DNA Nanostructures Using Relaxases". Angewandte Chemie International Edition. 55 (13): 4348–4352. doi:10.1002/anie.201510313. ISSN 1521-3773. PMC 5067690. PMID 26915475.
- Mie, Masayasu; Niimi, Takahiro; Mashimo, Yasumasa; Kobatake, Eiry (2019-01-03). "Construction of DNA-NanoLuc luciferase conjugates for DNA aptamer-based sandwich assay using Rep protein". Biotechnology Letters. 41 (3): 357–362. doi:10.1007/s10529-018-02641-7. ISSN 0141-5492. PMID 30603832.