Exonuclease 1
Exonuclease 1 is an enzyme that in humans is encoded by the EXO1 gene.[5][6][7]
This gene encodes a protein with 5' to 3' exonuclease activity as well as an RNase H activity (endonuclease activity cleaving RNA on DNA/RNA hybrid).[8] It is similar to the Saccharomyces cerevisiae protein Exo1 which interacts with Msh2 and which is involved in DNA mismatch repair and homologous recombination. Alternative splicing of this gene results in three transcript variants encoding two different isoforms.[7]
Meiosis
ExoI is essential for meiotic progression through metaphase I in the budding yeast Saccharomyces cerevisiae and in mouse.[9][10]
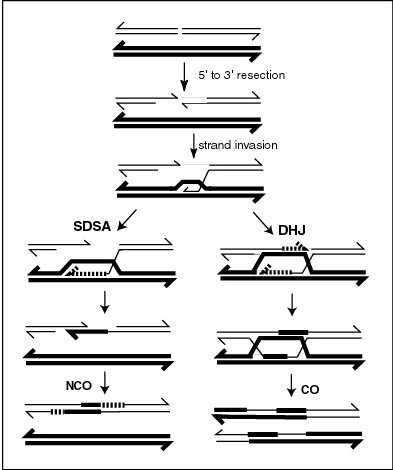
Recombination during meiosis is often initiated by a DNA double-strand break (DSB) as illustrated in the accompanying diagram. During recombination, sections of DNA at the 5' ends of the break are cut away in a process called resection. In the strand invasion step that follows, an overhanging 3' end of the broken DNA molecule "invades" the DNA of a homologous chromosome that is not broken, forming a displacement loop (D-loop). After strand invasion, the further sequence of events may follow either of two main pathways leading to a crossover (CO) or a non-crossover (NCO) recombinant (see Genetic recombination and Homologous recombination). The pathway leading to a CO involves a double Holliday junction (DHJ) intermediate. Holliday junctions need to be resolved for CO recombination to be completed.
During meiosis in S. cerevisiae, transcription of the Exo1 gene is highly induced.[9] In meiotic cells, Exo1 mutation reduces the processing of DSBs and the frequency of COs.[9] Exo1 has two temporally and biochemically distinct functions in meiotic recombination.[11] First, Exo1 acts as a 5’–3’ nuclease to resect DSB-ends. Later in the recombination process, Exo1 acts to facilitate the resolution of DHJs into COs, independently of its nuclease activities. In resolving DHJs, Exo 1 acts together with MLH1-MLH3 heterodimer (MutL gamma) and Sgs1 (ortholog of Bloom syndrome helicase) to define a joint molecule resolution pathway that produces the majority of crossovers.[12]
Male mice deficient for Exo1 are capable of normal progress through the pachynema stage of meiosis, but most germ cells fail to progress normally to metaphase I due to dynamic loss of chiasmata .[10]
Interactions
Exonuclease 1 has been shown to interact with MSH2[6][13][14] and MLH1.[14]
References
- GRCh38: Ensembl release 89: ENSG00000174371 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000039748 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Wilson DM III, Carney JP, Coleman MA, Adamson AW, Christensen M, Lamerdin JE (September 1998). "Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1". Nucleic Acids Res. 26 (16): 3762–8. doi:10.1093/nar/26.16.3762. PMC 147753. PMID 9685493.
- Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R (November 1998). "Human exonuclease I interacts with the mismatch repair protein hMSH2". Cancer Res. 58 (20): 4537–42. PMID 9788596.
- "Entrez Gene: EXO1 exonuclease 1".
- Qiu J, Qian Y, Chen V, Guan MX, Shen B (June 1999). "Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance". J. Biol. Chem. 274 (25): 17893–900. doi:10.1074/jbc.274.25.17893. PMID 10364235.
- Tsubouchi H, Ogawa H (2000). "Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae". Mol. Biol. Cell. 11 (7): 2221–33. doi:10.1091/mbc.11.7.2221. PMC 14915. PMID 10888664.
- Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Kneitz B, Yang G, Kunkel TA, Kolodner RD, Cohen PE, Edelmann W (2003). "Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility". Genes Dev. 17 (5): 603–14. doi:10.1101/gad.1060603. PMC 196005. PMID 12629043.
- Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N (2010). "Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions". Mol. Cell. 40 (6): 1001–15. doi:10.1016/j.molcel.2010.11.032. PMC 3061447. PMID 21172664.
- Zakharyevich K, Tang S, Ma Y, Hunter N (2012). "Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase". Cell. 149 (2): 334–47. doi:10.1016/j.cell.2012.03.023. PMC 3377385. PMID 22500800.
- Rasmussen, L J; Rasmussen M; Lee B; Rasmussen A K; Wilson D M; Nielsen F C; Bisgaard H C (June 2000). "Identification of factors interacting with hMSH2 in the fetal liver utilizing the yeast two-hybrid system. In vivo interaction through the C-terminal domains of hEXO1 and hMSH2 and comparative expression analysis". Mutat. Res. 460 (1): 41–52. CiteSeerX 10.1.1.614.1507. doi:10.1016/S0921-8777(00)00012-4. ISSN 0027-5107. PMID 10856833.
- Schmutte, C; Sadoff M M; Shim K S; Acharya S; Fishel R (August 2001). "The interaction of DNA mismatch repair proteins with human exonuclease I". J. Biol. Chem. 276 (35): 33011–8. doi:10.1074/jbc.M102670200. ISSN 0021-9258. PMID 11427529.
Further reading
- Liberti SE, Rasmussen LJ (2005). "Is hEXO1 a cancer predisposing gene?". Mol. Cancer Res. 2 (8): 427–32. PMID 15328369.
- Holle GE (1985). "[Pathophysiology of ulcer disease]". Langenbecks Archiv für Chirurgie. 366: 81–7. doi:10.1007/bf01836609. PMID 2414623.
- Bonaldo MF, Lennon G, Soares MB (1997). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Res. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Tishkoff DX, Amin NS, Viars CS, et al. (1998). "Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination". Cancer Res. 58 (22): 5027–31. PMID 9823303.
- Qiu J, Qian Y, Chen V, et al. (1999). "Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance". J. Biol. Chem. 274 (25): 17893–900. doi:10.1074/jbc.274.25.17893. PMID 10364235.
- Lee BI, Wilson DM (2000). "The RAD2 domain of human exonuclease 1 exhibits 5' to 3' exonuclease and flap structure-specific endonuclease activities". J. Biol. Chem. 274 (53): 37763–9. doi:10.1074/jbc.274.53.37763. PMID 10608837.
- Rasmussen LJ, Rasmussen M, Lee B, et al. (2000). "Identification of factors interacting with hMSH2 in the fetal liver utilizing the yeast two-hybrid system. In vivo interaction through the C-terminal domains of hEXO1 and hMSH2 and comparative expression analysis". Mutat. Res. 460 (1): 41–52. CiteSeerX 10.1.1.614.1507. doi:10.1016/S0921-8777(00)00012-4. PMID 10856833.
- Wu Y, Berends MJ, Post JG, et al. (2001). "Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms". Gastroenterology. 120 (7): 1580–7. doi:10.1053/gast.2001.25117. PMID 11375940.
- Schmutte C, Sadoff MM, Shim KS, et al. (2001). "The interaction of DNA mismatch repair proteins with human exonuclease I." J. Biol. Chem. 276 (35): 33011–8. doi:10.1074/jbc.M102670200. PMID 11427529.
- Jäger AC, Rasmussen M, Bisgaard HC, et al. (2001). "HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes". Oncogene. 20 (27): 3590–5. doi:10.1038/sj.onc.1204467. PMID 11429708.
- Genschel J, Bazemore LR, Modrich P (2002). "Human exonuclease I is required for 5' and 3' mismatch repair". J. Biol. Chem. 277 (15): 13302–11. doi:10.1074/jbc.M111854200. PMID 11809771.
- Lee Bi BI, Nguyen LH, Barsky D, et al. (2002). "Molecular interactions of human Exo1 with DNA". Nucleic Acids Res. 30 (4): 942–9. doi:10.1093/nar/30.4.942. PMC 100345. PMID 11842105.
- Sun X, Zheng L, Shen B (2002). "Functional alterations of human exonuclease 1 mutants identified in atypical hereditary nonpolyposis colorectal cancer syndrome". Cancer Res. 62 (21): 6026–30. PMID 12414623.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Jagmohan-Changur S, Poikonen T, Vilkki S, et al. (2003). "EXO1 variants occur commonly in normal population: evidence against a role in hereditary nonpolyposis colorectal cancer". Cancer Res. 63 (1): 154–8. PMID 12517792.
- Sharma S, Sommers JA, Driscoll HC, et al. (2003). "The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein". J. Biol. Chem. 278 (26): 23487–96. doi:10.1074/jbc.M212798200. PMID 12704184.
- Alam NA, Gorman P, Jaeger EE, et al. (2004). "Germline deletions of EXO1 do not cause colorectal tumors and lesions which are null for EXO1 do not have microsatellite instability". Cancer Genet. Cytogenet. 147 (2): 121–7. doi:10.1016/S0165-4608(03)00196-1. PMID 14623461.
- Genschel J, Modrich P (2004). "Mechanism of 5'-directed excision in human mismatch repair". Mol. Cell. 12 (5): 1077–86. doi:10.1016/S1097-2765(03)00428-3. PMID 14636568.