ELMO (protein)
ELMO (Engulfment and Cell Motility) is a family of related proteins (~82 kDa) involved in intracellular signalling networks. These proteins have no intrinsic catalytic activity and instead function as adaptors which can regulate the activity of other proteins through their ability to mediate protein-protein interactions.
| ELMO/CED-12 family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ELMO_CED12 | ||||||||
| Pfam | PF04727 | ||||||||
| InterPro | IPR006816 | ||||||||
| PROSITE | PDOC51335 | ||||||||
| |||||||||
This family contains members in all animals. In humans there are three paralogous isoforms:
The ELMO domain was first characterized in the CED-12 proteins of Caenorhabditis elegans and Drosophila melanogaster. CED-12 is a homolog to the ELMO1 protein found in mammals. This protein is involved in Rac-GTPase activation, apoptotic cell phagocytosis, cell migration, and cytoskeletal rearrangements.[1][2]
Discovery of CED-12
The discovery of CED-12 was done using knockout experiments.[1] Its involvement in the apoptotic phagocytosis pathway was first noted when knocked-out ced-12 in C. elegans showed similar results in the apoptotic process to ced-5 and ced-2 knockouts.[3] This lead researchers to believe, and later confirm, that the protein products of ced-12 (CED-12), ced-5 (CED-5), and ced-2 (CED-2) all functioned as part of the same pathway.[3][4]
Researchers also noted direct protein-protein interactions between CED-12 and CED-10 (C. elegans homolog for Rac1), a Rac-GTPase (energy-dependent protein found used for cytoskeletal rearrangements among other functions).[5][6] CED-10 was inactive when CED-12 was knocked-out. Expression of CED-12 with CED-5 and CED-2 activated CED-10, which lead to the activation of apoptotic phagocytosis.[3]
Function of CED-12
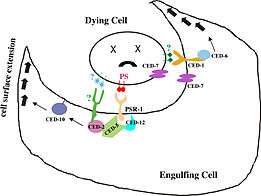
CED-12 is an adaptor protein (proteins involved in facilitating the formation of signalling complexes) that is translated once apoptosis has been triggered in a cell. Apoptosis, also known as programmed cell death, activates during development as well as in situations where a cell has received sufficient physical damage.[7][8] Many of the contents within a cell are reactive with the environment outside of the cell and must be disposed of without causing any harm to the surrounding tissues. Apoptotic cells are removed from their external environment by neighbouring cells that recognize cell-surface markers located on the apoptotic cell membrane. Marker recognition leads to the engulfment of apoptotic cells by phagocytosis.[8] On a molecular level, recognition of the cell-surface markers leads to the translation of the CED-12 protein in the cytoplasm of the engulfing cell, which then gets localized to the cell membrane. CED-12 binds CED-2 (C. elegans homolog to CrkII in mammals), followed by CED-5 (C. elegans homolog for DOCK180 in mammals) and forms a ternary structure.[5][9] Transmembrane CED-1 is an example of the cell-surface receptor on the engulfing cell. When receptors come in contact with cell surface markers on the apoptotic cell, a protein known as CED-6 (homolog for GULP in mammals) is expressed.[2][10] Both the CED-2/CED-5/CED-12 ternary structure and CED-6 function to activate an effector protein known as CED-10. CED-10 is a RAC-GTPase protein that is directly responsible for the rearrangement of the actin cytoskeleton that initiates phagocytosis.[5][6] This process is regulated by two pathways. The first is by CED-6, which is an adaptor protein that is responsible for coordinating protein-protein interactions between CED-10 and actin.[11] The second pathway occurs when the CED-2/CED-5/CED-12 ternary structure form a GEF (guanine nucleotide exchange factor) with CED-10, which promotes the binding of a GTP energy molecule in order to activate the GTP-dependent CED-10.[2][5][10][11]
CED-12 also functions in cell migration processes, which is regulated by the same interactions as the apoptotic phagocytosis pathway. It functions in distal tip cell migration in gonad development in C. elegans.[12] Distal tip cells are somatic cells located at the tip of developing gonadal arms, and are responsible for the elongation of the gonadal arm as well as controlling mitotic and meiotic cell division of gonadal cells throughout development and adulthood.[13] As C. elegans develops, the distal cells undergo a series of migrations in order to complete morphological changes, which define both gonad shape and size.[12] This process occurs when integrins on the surface of the distal tip cells meet chemoattractants located on the extracellular matrix.[12][13] The integrins form focal adhesions at the sites of the chemoattractants, which causes the localization of CED-5 to the adhesion points.[12] CED-12 and CED-2 form the GEF-trio with CED-5 and activate the CED-10 Rac-GTPase in order to rearrange the actin cytoskeleton and promote the forward propagation of the distal tip cells.[12][14]
Gene and Protein Structure of CED-12
_and_DOCK2_(CED-5)_ternary_complex_in_apoptosis.jpg)
The ced-12 gene codes for an 82kDa large protein, which spans 731 amino acids in length.[2] It is found on chromosome 2 on the L-arm in Drosophila, and on chromosome I in C. elegans.[1] The protein structure of CED-12 is separated based on its binding domains:
- The proline-rich region on CED-12 is a binding site for the C-terminal SH3-binding domain on CED-5/DOCK180.[15] The proline-rich region contains a high concentration of the amino acid Proline, and lies between amino acid residues 711–724.[2] This domain is crucial in the remodelling processes of the cytoskeleton and follows a conserved sequence pattern composed of proline and arbitrary aliphatic (non-polar amino acids with open alkane side chains) residues.[2] The conserved pattern of the sequence allows for hydrophobic and salt-bridge interactions with the SH3-domain.[15]
- The repeating Armadillo (ARM) region on the N-terminal binds CED-2/CrkII, which is necessary to activate the heterodimerization with CED-5/DOCK180.[11]
- The Pleckstrin Homology domain spans 100-200 amino acids in length.[2][11][16] It is located close to the C-terminal and is necessary to bind the Rac-GTPase once the Guanine Nucleotide Exchange Factor with CED-5 and CED-2 is formed. This activates the cytoskeletal remodelling.[11]
Structure and function of ELMO proteins
The ELMO family are evolutionarily conserved orthologs of the C. elegans protein CED-12. All isoforms contain a series of armadillo repeats, which begin at the N-terminus and extend around two thirds of the way along the protein, as well as a C-terminal proline-rich motif and a central PH domain.[17] They function as part of a protein complex with Dock180-related proteins to form a bipartite guanine nucleotide exchange factor for Rac (a member of the Rho family of small G proteins).[18] The Dock180-ELMO interaction requires the ELMO PH domain and also involves binding of the ELMO proline-rich motif to the Dock180 SH3 domain.[19]
References
- Brody, Thomas. "Ced-12". The Interactive Fly. Retrieved November 11, 2015.
- Zhou Z, Caron E, Hartwieg E, Hall A, Horvitz HR (October 2001). "The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway". Developmental Cell. 1 (4): 477–89. doi:10.1016/s1534-5807(01)00058-2. PMID 11703939.
- Pasqualini, Renata; Arap, Wadih (2009). Protein Discovery Technologies. CRC Press. p. 175. ISBN 978-1420014211.
- Chung S, Gumienny TL, Hengartner MO, Driscoll M (December 2000). "A common set of engulfment genes mediates removal of both apoptotic and necrotic cell corpses in C. elegans". Nature Cell Biology. 2 (12): 931–7. doi:10.1038/35046585. PMID 11146658.
- Lettre G, Hengartner MO (February 2006). "Developmental apoptosis in C. elegans: a complex CEDnario". Nature Reviews. Molecular Cell Biology. 7 (2): 97–108. doi:10.1038/nrm1836. PMID 16493416.
- Raftopoulou M, Hall A (January 2004). "Cell migration: Rho GTPases lead the way". Developmental Biology. 265 (1): 23–32. doi:10.1016/j.ydbio.2003.06.003. PMID 14697350.
- Programmed cell death. www.ncbi.nlm.nih.gov. WormBook. 2005-10-06. Retrieved 2015-12-02.
- Elmore S (June 2007). "Apoptosis: a review of programmed cell death". Toxicologic Pathology. 35 (4): 495–516. doi:10.1080/01926230701320337. PMC 2117903. PMID 17562483.
- Wang X, Wu YC, Fadok VA, Lee MC, Gengyo-Ando K, Cheng LC, et al. (November 2003). "Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12". Science. 302 (5650): 1563–6. Bibcode:2003Sci...302.1563W. doi:10.1126/science.1087641. PMID 14645848.
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, et al. (March 2005). "Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans". Nature. 434 (7029): 93–9. Bibcode:2005Natur.434...93K. doi:10.1038/nature03263. PMID 15744306.
- Ravichandran KS, Lorenz U (December 2007). "Engulfment of apoptotic cells: signals for a good meal". Nature Reviews. Immunology. 7 (12): 964–74. doi:10.1038/nri2214. PMID 18037898.
- Wong MC, Schwarzbauer JE (2012). "Gonad morphogenesis and distal tip cell migration in the Caenorhabditis elegans hermaphrodite". Wiley Interdisciplinary Reviews. Developmental Biology. 1 (4): 519–31. doi:10.1002/wdev.45. PMC 3614366. PMID 23559979.
- "Reproductive System: The Somatic Gonad".
- Conradt B (October 2001). "Cell engulfment, no sooner ced than done". Developmental Cell. 1 (4): 445–7. doi:10.1016/s1534-5807(01)00065-x. PMID 11703934.
- Weng Z, Rickles RJ, Feng S, Richard S, Shaw AS, Schreiber SL, Brugge JS (October 1995). "Structure-function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions". Molecular and Cellular Biology. 15 (10): 5627–34. doi:10.1128/mcb.15.10.5627. PMC 230813. PMID 7565714.
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, et al. (October 2001). "CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration" (PDF). Cell. 107 (1): 27–41. doi:10.1016/s0092-8674(01)00520-7. PMID 11595183.
- Lu M, Ravichandran KS (2006). "Dock180-ELMO cooperation in Rac activation". Methods in Enzymology. 406: 388–402. doi:10.1016/S0076-6879(06)06028-9. ISBN 9780121828110. PMID 16472672.
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, et al. (October 2001). "CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration" (PDF). Cell. 107 (1): 27–41. doi:10.1016/S0092-8674(01)00520-7. PMID 11595183.
- Komander D, Patel M, Laurin M, Fradet N, Pelletier A, Barford D, Côté JF (November 2008). "An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling". Molecular Biology of the Cell. 19 (11): 4837–51. doi:10.1091/mbc.E08-04-0345. PMC 2575150. PMID 18768751.