Drosophila circadian rhythm
Drosophila circadian rhythm is a daily 24-hour cycle of rest and activity in the fruit flies of the genus Drosophila. The biological process was discovered and is best understood in the species Drosophila melanogaster. Other than normal sleep-wake activity, D. melanogaster has two unique daily behaviours, namely regular vibration (locomotor activity) during the process of hatching (called eclosion) from the pupa, and during mating. Locomotor activity is maximum at dawn and dusk, while eclosion is at dawn.[1]
.jpg)
Biological rhythms were first studied in Drosophila. Drosophila circadian rhythm have paved the way for understanding circadian behaviour and diseases related to sleep-wake conditions in other animals, including humans. This is because the circadian clocks are fundamentally similar.[2] Drosophila circadian rhythm was discovered in 1935 by German zoologists, Hans Kalmus and Erwin Bünning. American biologist Colin S. Pittendrigh provided an important experiment in 1954, which established that circadian rhythm is driven by a biological clock. The genetics was first understood in 1971, when Seymour Benzer and Ronald J. Konopka reported that mutation in specific genes changes or stops the circadian behaviour. They discovered the gene called period (per), mutations of which alter the circadian rhythm. It was the first gene known to control behaviour. After a decade, Konopka, Jeffrey C. Hall, Michael Rosbash, and Michael W. Young discovered novel genes including timeless (tim), Clock (Clk), cycle (cyc), cry. These genes and their product proteins play a key role in the circadian clock.
For their contributions, Hall, Rosbash and Young received the Nobel Prize in Physiology or Medicine in 2017.[3]
History
During the process of eclosion by which an adult fly emerges from the pupa, Drosophila exhibits regular locomotor activity (by vibration) that occurs during 8-10 hours intervals starting just before dawn. The existence of this circadian rhythm was independently discovered in D. melanogaster in 1935 by two German zoologists, Hans Kalmus at the Zoological Institute of the German University in Prague (now Charles University), and Erwin Bünning at the Botanical Institute of the University of Jena.[4][5] Kalmus discovered in 1938 that the brain area is responsible for the circadian activity.[6] Kalmus and Bünning were of the opinion that temperature was the main factor. But it was soon realized that even in different temperature, the circadian rhythm could be unchanged.[7] In 1954, Colin S. Pittendrigh at the Princeton University discovered the importance of light-dark conditions in D. pseudoobscura. He demonstrated that eclosion rhythm was delayed but not stopped when temperature was decreased. He concluded that temperature influenced only the peak hour of the rhythm, and was not the principal factor.[8] It was then known that the circadian rhythm was controlled by a biological clock. But the nature of the clock was then a mystery.[5]
After almost two decades, the existence of the circadian clock was discovered by Seymour Benzer and his student Ronald J. Konopka at the California Institute of Technology. They discovered that mutations in the X chromosome of D. melanogaster could make abnormal circadian activities. When a specific part of the chromosome was absent (inactivated), there was no circadian rhythm; in one mutation (called perS, "S" for short or shortened) the rhythm was shortened to ~19 hours; whereas, in another mutation (perL, "L" for long or lengthened) the rhythm was extended to ~29 hours, as opposed to a normal 24 hour rhythm. They published the discovery in 1971.[9] They named the gene location (locus) as period (per for short) as it controls the period of the rhythm. In opposition, there were other scientists that stated genes could not control such complex behaviors as circadian activities.[10]
Another circadian behavior in Drosophila is courtship between the male and female during mating. Courtship involves a song accompanied by a ritual locomotory dance in males. The main flight activity generally takes place in the morning and another peak occurs before sunset. Courtship song is produced by the male's wing vibration and consists of pulses of tone produced at intervals of approximately 34 msec in D. melanogaster (48 msec in D. simulans). In 1980, Jeffrey C. Hall and his student Charalambos P. Kyriacou, at Brandeis University in Waltham, discovered that courtship activity is also controlled by per gene.[11] In 1984, Konopka, Hall, Michael Roshbash and their team reported in two papers that per locus is the centre of the circadian rhythm, and that loss of per stops circadian activity.[12][13] At the same time, Michael W. Young's team at the Rockefeller University reported similar effects of per, and that the gene covers 7.1-kilobase (kb) interval on the X chromosome and encodes a 4.5-kb poly(A)+ RNA.[14][15] In 1986, they sequenced the entire DNA fragment and found the gene encodes the 4.5-kb RNA, which produces a protein, a proteoglycan, composed of 1,127 amino acids.[16] At the same time Roshbash's team showed that PER protein is absent in mutant per.[17] In 1994, Young and his team discovered the gene timeless (tim) that influences the activity of per.[18] In 1998, they discovered doubletime (dbt), which regulate the amount of PER protein.[19]
In 1990, Konopka, Rosbash, and identified a new gene called Clock (Clk), which is vital for the circadian period.[20] In 1998, they found a new gene cycle (cyc), which act together with Clk.[21] In the late 1998, Hall and Roshbash's team discovered crybaby, a gene for sensitivity to blue light.[22] They simultaneously identified the protein CRY as the main light-sensitive (photoreceptor) system. The activity of cry is under circadian regulation, and influenced by other genes such as per, tim, clk, and cyc.[23] The gene product CRY is a major photoreceptor protein belonging to a class of flavoproteins called cryptochromes. They are also present in bacteria and plants.[24] In 1998, Hall and Jae H. Park isolated a gene encoding a neuropeptide named pigment dispersing factor (PDF), based on one of the roles it plays in crustaceans.[25] In 1999, they discovered that pdf is expressed by lateral neurone ventral clusters (LNv) indicating that PDF protein is the major circadian neurotransmitter and that the LNv neurones are the principal circadian pacemakers.[26] In 2001, Young and his team demonstrated that glycogen synthase kinase-3 (GSK-3) ortholog shaggy (SGG) is an enzyme that regulates TIM maturation and accumulation in the early night, by causing phosphorylation.[27]
Hall, Rosbash and Young shared the Nobel Prize in Physiology or Medicine 2017 “for their discoveries of molecular mechanisms controlling the circadian rhythm”.[3]
Mechanism

In Drosophila there are two distinct groups of circadian clocks, namely the clock neurones and the clock genes. They act concertedly to produce the 24-hour cycle of rest and activity. Light is the source of activation of the clocks. The compound eyes, ocelli, and Hofbauer-Buchner eyelets (HB eyelets) are the direct external photoreceptor organs. But the circadian clock can work in constant darkness.[28] Nonetheless, the photoreceptors are required for measuring the day length and detecting moonlight. The compound eyes are important for differentiating long days from constant light, and for the normal masking effects of light, such as inducing activity by light and inhibition by darkness.[29] There are two distinct activity peaks termed the M (for morning) peak, happening at dawn, and E (for evening) peak, at dusk. They monitor the different day lengths in different seasons of the year.[30] The light-sensitive proteins in the eye called, rhodopsins (rhodopsin 1 and 6), are crucial in activating the M and E oscillations.[31] When environmental light is detected, approximately 150 neurones (there are about 100,000 neurones in the Drosophila brain) in the brain regulate the circadian rhythm.[32] The clock neurones are located in distinct clusters in the central brain. The best-understood clock neurones are the large and small lateral ventral neurons (l-LNvs and s-LNvs) of the optic lobe. These neurones produce pigment dispersing factor (PDF), a neuropeptide that acts as a circadian neuromodulator between different clock neurones.[33]
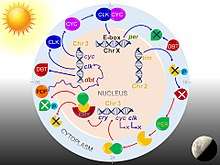
Drosophila circadian keeps time via daily fluctuations of clock-related proteins which interact in what is called a transcription-translation feedback loop. The core clock mechanism consists of two interdependent feedback loops, namely the PER/TIM loop and the CLK/CYC loop.[34] The CLK/CYC loop occurs during the day in which both Clock protein and cycle protein are produced. CLK/CYC heterodimer act as transcription factors and bind together to initiate the transcription of the per and tim genes by binding to a promoter element called E box, around mid-day. DNA is transcribed to produce PER mRNA and TIM mRNA. PER and TIM proteins are synthesized in the cytoplasm, and exhibit a smooth increase in levels over the course of the day. Their RNA levels peak early in the evening and protein levels peak around daybreak.[32] But their proteins levels are maintained at constantly low level until dusk, because during daylight also activates the doubletime (dbt) gene. DBT protein induces post-translational modifications, that is phosphorylation and turnover of monomeric PER proteins. As PER is translated in the cytoplasm, it is actively phosphorylated by DBT (casein kinase 1ε) and casein kinase 2 (synthesised by And and Tik) as a prelude to premature degradation. The actual degradation is through the ubiquitin-proteasome pathway, and is carried out by a ubiquitin ligase called Slimb (supernumery limbs).[35][36] At the same time, TIM is itself phosphorylated by shaggy, whose activity declines after sunset. DBT gradually disappears, and withdrawal of DBT promotes PER molecules to get stabilized by physical association with TIM. Hence, maximum production of PER and TIM occurs at dusk. At the same time, CLK/CYC also directly activates vri and Pdp1 (the gene for PAR domain protein 1). VRI accumulates first, 3-6 hour earlier, and start to repress Clk; but the incoming of PDP1 creates a competition by activating Clk. PER/TIM dimer accumulate in the early night and translocate in an orchestrated fashion into the nucleus several hours later, and binds to CLK/CYC dimers. Bound PER completely stops the transcriptional activity of CLK and CYC.[37]
In the early morning, the appearance of light causes PER and TIM proteins to breakdown in a network of transcriptional activation and repression. First, light activates the cry gene in the clock neurones. Although CRY is produced deep inside the brain, it is sensitive to UV and blue light, and thus it easily signals the brain cells the onset of light. It irreversibly and directly binds to TIM causing it to break down through proteosome-dependent ubiquitin-mediated degradation. The CRY's photolyase homology domain is used for light detection and phototransduction, whereas the carboxyl-terminal domain regulates CRY stability, CRY-TIM interaction, and circadian photosensitivity.[38] The ubiquitination and subsequent degradation are aided by a different protein JET.[39] Thus PER/TIM dimer dissociates, and the unbound PER becomes unstable. PER undergoes progressive phosphorylation and ultimately degradation. Absence of PER and TIM allows activation of clk and cyc genes. Thus, the clock is reset to commence the next circadian cycle.[10]
References
- Dubowy, Christine; Sehgal, Amita (2017). "Circadian Rhythms and Sleep in". Genetics. 205 (4): 1373–1397. doi:10.1534/genetics.115.185157. PMC 5378101. PMID 28360128.
- Rosato, Ezio; Tauber, Eran; Kyriacou, Charalambos P (2006). "Molecular genetics of the fruit-fly circadian clock". European Journal of Human Genetics. 14 (6): 729–738. doi:10.1038/sj.ejhg.5201547. PMID 16721409.
- Nobel Foundation (2017). "The Nobel Prize in Physiology or Medicine 2017". www.nobelprize.org. Nobel Media AB. Retrieved 28 December 2017.
- Bruce, Victor G.; Pittendrigh, Colin S. (1957). "Endogenous Rhythms in Insects and Microorganisms". The American Naturalist. 91 (858): 179–195. doi:10.1086/281977.
- Pittendrigh, C. S. (1993). "Temporal Organization: Reflections of a Darwinian Clock-Watcher". Annual Review of Physiology. 55 (1): 17–54. doi:10.1146/annurev.ph.55.030193.000313. PMID 8466172.
- Kalmus, H. (1938). "Die Lage des Aufnahmeorganes für die Schlupfperiodik von Drosophila [The location of the receiving organ for the hatching period of Drosophila]". Zeitschrift für vergleichende Physiologie. 26 (3): 362–365. doi:10.1007/BF00338939.
- Welsh, J.H. (1938). "Diurnal rhythms". The Quarterly Review of Biology. 13 (2): 123–139. doi:10.1086/394554.
- Pittendrigh, C.S. (1954). "On temperature independence in the clock system controlling emergence time in Drosophila". Proceedings of the National Academy of Sciences of the United States of America. 40 (10): 1018–1029. doi:10.1073/pnas.40.10.1018. PMC 534216. PMID 16589583.
- Konopka, R.J.; Benzer, S. (1971). "Clock mutants of Drosophila melanogaster". Proceedings of the National Academy of Sciences of the United States of America. 68 (9): 2112–2116. doi:10.1073/pnas.68.9.2112. PMC 389363. PMID 5002428.
- Lalchhandama, K. (2017). "The path to the 2017 Nobel Prize in Physiology or Medicine". Science Vision. 3 (Suppl): 1–13.
- Kyriacou, C.P.; Hall, J.C. (1980). "Circadian rhythm mutations in Drosophila melanogaster affect short-term fluctuations in the male's courtship song". Proceedings of the National Academy of Sciences of the United States of America. 77 (11): 6729–6733. doi:10.1073/pnas.77.11.6729. PMC 350362. PMID 6779281.
- Reddy, P.; Zehring, W.A.; Wheeler, D.A.; Pirrotta, V.; Hadfield, C.; Hall, J.C.; Rosbash, M. (1984). "Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms". Cell. 38 (3): 701–710. doi:10.1016/0092-8674(84)90265-4. PMID 6435882.
- Zehring, W.A.; Wheeler, D.A.; Reddy, P.; Konopka, R.J.; Kyriacou, C.P.; Rosbash, M.; Hall, J.C. (1984). "P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster". Cell. 39 (2 Pt 1): 369–376. doi:10.1016/0092-8674(84)90015-1. PMID 6094014.
- Bargiello, T.A.; Jackson, F.R.; Young, M.W. (1984). "Restoration of circadian behavioural rhythms by gene transfer in Drosophila". Nature. 312 (5996): 752–754. doi:10.1038/312752a0. PMID 6440029.
- Bargiello, T.A.; Young, M.W. (1984). "Molecular genetics of a biological clock in Drosophila". Proceedings of the National Academy of Sciences of the United States of America. 81 (7): 2142–2146. doi:10.1038/312752a0. PMC 345453. PMID 16593450.
- Jackson, F.R.; Bargiello, T.A.; Yun, S.H.; Young, M.W. (1986). "Product of per locus of Drosophila shares homology with proteoglycans". Nature. 320 (6058): 185–188. doi:10.1038/320185a0. PMID 3081818.
- Reddy, P.; Jacquier, A.C.; Abovich, N.; Petersen, G.; Rosbash, M. (1986). "The period clock locus of D. melanogaster codes for a proteoglycan". Cell. 46 (1): 53–61. doi:10.1016/0092-8674(86)90859-7. PMID 3087625.
- Sehgal, A.; Price, J.L.; Man, B.; Young, M.W. (1994). "Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless". Science. 263 (5153): 1603–1606. doi:10.1126/science.8128246. PMID 8128246.
- Price, J.L.; Blau, J.; Rothenfluh, A.; Abodeely, M.; Kloss, B.; Young, M.W. (1998). "double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation". Cell. 94 (1): 83–95. doi:10.1016/S0092-8674(00)81224-6. PMID 9674430.
- Dushay, M.S.; Konopka, R.J.; Orr, D.; Greenacre, M.L.; Kyriacou, C.P.; Rosbash, M.; Hall, J.C. (1990). "Phenotypic and genetic analysis of Clock, a new circadian rhythm mutant in Drosophila melanogaster". Genetics. 125 (3): 557–578. PMC 1204083. PMID 2116357.
- Rutila, J.E.; Suri, V.; Le, M.; So, W.V.; Rosbash, M.; Hall, J.C. (1998). "CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless". Cell. 93 (5): 805–814. doi:10.1016/S0092-8674(00)81441-5. PMID 9630224.
- Stanewsky, R.; Kaneko, M.; Emery, P.; Beretta, B.; Wager-Smith, K.; Kay, S.A.; Rosbash, M.; Hall, J.C. (1998). "The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila". Cell. 95 (5): 681–692. doi:10.1016/s0092-8674(00)81638-4. PMID 9845370.
- Emery, P.; So, W.V.; Kaneko, M.; Hall, J.C.; Rosbash, M. (1998). "CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity". Cell. 95 (5): 669–679. doi:10.1016/S0092-8674(00)81637-2. PMID 9845369.
- Mei, Q.; Dvornyk, V. (2015). "Evolutionary History of the Photolyase/Cryptochrome Superfamily in Eukaryotes". PLOS One. 10 (9): e0135940. doi:10.1371/journal.pone.0135940. PMC 4564169. PMID 26352435.
- Park, J.H.; Hall, J.C. (1998). "Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster". Journal of Biological Rhythms. 13 (3): 219–228. doi:10.1177/074873098129000066. PMID 9615286.
- Renn, S.C.; Park, J.H.; Rosbash, M.; Hall, J.C.; Taghert, P.H. (1999). "A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila". Cell. 99 (7): 791–802. doi:10.1016/s0092-8674(00)81676-1. PMID 10619432.
- Martinek, S.; Inonog, S.; Manoukian, A.S.; Young, M.W. (2001). "A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock". Cell. 105 (6): 769–779. doi:10.1016/S0092-8674(01)00383-X. PMID 11440719.
- Veleri, S.; Wülbeck, C. (2004). "Unique self-sustaining circadian oscillators within the brain of Drosophila melanogaster". Chronobiology International. 21 (3): 329–342. doi:10.1081/CBI-120038597. PMID 15332440.
- Rieger, D.; Stanewsky, R.; Helfrich-Förster, C. (2003). "Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster". Journal of Biological Rhythms. 18 (5): 377–391. doi:10.1177/0748730403256997. PMID 14582854.
- Yoshii, T.; Rieger, D.; Helfrich-Förster, C. (2012). "Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila". Progress in Brain Research. 199 (1): 59–82. doi:10.1016/B978-0-444-59427-3.00027-7. PMID 22877659.
- Schlichting, M.; Grebler, R.; Peschel, N.; Yoshii, T.; Helfrich-Förster, C. (2014). "Moonlight detection by Drosophila's endogenous clock depends on multiple photopigments in the compound eyes". Journal of Biological Rhythms. 29 (2): 75–86. doi:10.1177/0748730413520428. PMID 24682202.
- Nitabach, M.N.; Taghert, P.H. (2008). "Organization of the Drosophila circadian control circuit". Current Biology. 18 (2): 84–93. doi:10.1016/j.cub.2007.11.061. PMID 18211849.
- Yoshii, T.; Hermann-Luibl, C.; Helfrich-Förster, C. (2015). "Circadian light-input pathways in Drosophila". Communicative & Integrative Biology. 9 (1): e1102805. doi:10.1080/19420889.2015.1102805. PMC 4802797. PMID 27066180.
- Boothroyd, C.E.; Young, M.W. (2008). "The in(put)s and out(put)s of the Drosophila circadian clock". Annals of the New York Academy of Sciences. 1129: 350–357. doi:10.1196/annals.1417.006. PMID 18591494.
- Grima, B.; Lamouroux, A.; Chélot, E.; Papin, C.; Limbourg-Bouchon, B.; Rouyer, F. (2002). "The F-box protein slimb controls the levels of clock proteins period and timeless". Nature. 420 (6912): 178–182. doi:10.1038/nature01122. PMID 12432393.
- Ko, H.W.; Jiang, J.; Edery, I. (2002). "Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime". Nature. 420 (6916): 673–678. doi:10.1038/nature01272. PMID 12442174.
- Helfrich-Förster, C. (2005). "Neurobiology of the fruit fly's circadian clock". Genes, Brain, and Behavior. 4 (2): 65–76. doi:10.1111/j.1601-183X.2004.00092.x. PMID 15720403.
- Busza, A.; Emery-Le, M.; Rosbash, M.; Emery, P. (2004). "Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception". Science. 304 (5676): 1503–1506. doi:10.1126/science.1096973. PMID 15178801.
- Koh, K.; Zheng, X.; Sehgal, A. (2006). "JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS". Science. 312 (5781): 1809–1812. doi:10.1126/science.1124951. PMC 2767177. PMID 16794082.