Diauxie
Diauxie is a Greek word coined by Jacques Monod to mean two growth phases. The word is used in English in cell biology to describe the growth phases of a microorganism in batch culture as it metabolizes a mixture of two sugars. Rather than metabolizing the two available sugars simultaneously, microbial cells commonly consume them in a sequential pattern, resulting in two separate growth phases.
Growth phases
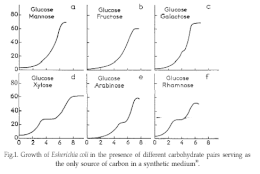
Jacques Monod discovered diauxic growth in 1941 during his experiments with Escherichia coli and Bacillus subtilis. While growing these bacteria on various combination of sugars during his doctoral thesis research, Monod observed that often two distinct growth phases are clearly visible in batch culture, as seen in Figure 1.
During the first phase, cells preferentially metabolize the sugar on which it can grow faster (often glucose but not always). Only after the first sugar has been exhausted do the cells switch to the second. At the time of the "diauxic shift", there is often a lag period during which cells produce the enzymes needed to metabolize the second sugar.
Monod later put aside his work on diauxic growth and focused on the lac operon model of gene expression, which led to a Nobel prize.
Diauxie occurs because organisms use operons or multiple sets of genes to control differently the expression of enzymes needed to metabolize the different nutrients or sugars they encounter. If an organism allocates its energy and other resources (e.g. amino acids) to synthesize enzymes needed to metabolize a sugar that can only support a slower growth rate and not use all or most of its available resources to synthesize the enzymes that metabolize a different sugar providing a faster growth rate, such an organism will be at a reproductive disadvantage compared to those that choose to grow on the faster growth supporting sugar. Through evolution, organisms have developed the ability to regulate their genetic control mechanisms so as to only express those genes resulting in the fastest growth rate. For example, when grown in the presence of both glucose and maltose, Lactococcus lactis will produce enzymes to metabolize glucose first, altering its gene expression to use maltose only after the supply of glucose has been exhausted.
Aerobic fermentation
In the case of the baker's or brewer's yeast Saccharomyces cerevisiae growing on glucose with plenty of aeration, the diauxic growth pattern is commonly observed in batch culture. During the first growth phase, when there is plenty of glucose and oxygen available, the yeast cells prefer glucose fermentation to aerobic respiration, in a phenomenon known as aerobic fermentation. Although aerobic respiration may seem a more energetically-efficient pathway to grow on glucose, it is in fact a rather inefficient way to increase biomass as most of the carbon in the glucose is oxidized to carbon dioxide rather than incorporated into new amino acids or fatty acids. Contrary to the more commonly invoked Pasteur effect, this phenomenon is closer to the Warburg effect observed in faster growing tumors.
The intracellular genetic regulatory mechanisms have evolved to enforce this choice, as fermentation provides a faster anabolic growth rate for the yeast cells than the aerobic respiration of glucose, which favors catabolism. After glucose is depleted, the fermentative product ethanol is oxidised in a noticeably slower second growth phase, if oxygen is available.
Proposed mechanisms
In the 1940s, Monod hypothesized that a single enzyme could adapt to metabolize different sugars. It took 15 years of further work to show that this was incorrect. During his work on the lac operon of E. coli, Joshua Lederberg isolated β-galactosidase and found it in greater quantities in colonies grown on lactose compared to other sugars. Melvin Cohn in Monod's lab at the Pasteur Institute then found that β-galactosides induced enzyme activity. The idea of enzyme adaptation was thus replaced with the concept of enzyme induction, in which a molecule induces expression of a gene or operon, often by binding to a repressor protein and preventing it from attaching to the operator.[1]
In the case of the bacterial diauxic shift from glucose to lactose metabolism, a proposed mechanism suggested that glucose initially inhibits the ability of the enzyme adenylate cyclase to synthesize cyclic AMP (cAMP). cAMP, in turn, is required for the catabolite activator protein (CAP) to bind to DNA and activate the transcription of the lac operon, which includes genes necessary for lactose metabolism. The presence of allolactose, a metabolic product of lactose, is sensed through the activity of the lac repressor, which inhibits transcription of the lac operon until lactose is present. Thus, if glucose is present, cAMP levels remain low, so CAP is unable to activate transcription of the lac operon, regardless of the presence or absence of lactose. Upon the exhaustion of the glucose supply, cAMP levels rise, allowing CAP to activate the genes necessary for the metabolism of other food sources, including lactose if it is present.[2]
More recent research however suggests that the cAMP model is not correct in this instance since cAMP levels remain identical under glucose and lactose growth conditions, and a different model has been proposed and it suggests that the lactose-glucose diauxie in E. coli may be caused mainly by inducer exclusion.[3] In this model, glucose transport via the EIIAGlc shuts down lactose permease when glucose is being transported into the cell, so lactose is not transported into cell and used. While the cAMP/CAP mechanism may not play a role in the glucose/lactose diauxie, it is a suggested mechanism for other diauxie .
References
- Mulligan, Martin. "Induction". Archived from the original on 2007-11-16. Retrieved 2007-01-01.
- Brown, T.A. "Transient Changes in Gene Activity". Retrieved 2007-01-01.
- Stülke J, Hillen W. (1999). "Carbon catabolite repression in bacteria". Current Opinion in Microbiology. 2 (2): 195–201. doi:10.1016/S1369-5274(99)80034-4. PMID 10322165.