Coiled-coil domain containing 42B
Coiled Coil Domain Containing protein 42B, also known as CCDC42B, is a protein encoded by the protein-coding gene CCDC42B.[5]
| CFAP73 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | CFAP73, MIA2, CCDC42B, Coiled-coil domain containing 42B, cilia and flagella associated protein 73 | ||||||||||||||||||||||||
| External IDs | MGI: 3779542 HomoloGene: 53205 GeneCards: CFAP73 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
| Location (UCSC) | Chr 12: 113.15 – 113.16 Mb | Chr 5: 120.63 – 120.63 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Locus
CCDC42B gene is located on the plus strand of chromosome12 at position 24.13 of the long arm. CCDC42B gene starts at 113,587,663 base pairs and end at 113,597,081 base pairs. Part of CCDC42B overlaps with DDX54 gene (113,594,978-113,623,284). The size of CCDC42B is 9,419 bases and its molecular weight is 35,914 Da.[5][6][7] CCDC42B mRNA contains 1514 bp and located from 113,587,663 to 113,597,081. CCDC42B protein contains 308 AA and located from 113,587,663 to 113,595,484. The promoter region (GXP_642107) contains 859 bp is predicted to be located from 113,586,906 to 113,587,764. Human CCDC42B gene has three neighbor gene: DDX54, RASAL 1,and DTX1.
DDX54 gene is member of DEAD protein family of Putative RNA helicases. The gene encodes DEAD box Protein which has a conserved motif of Asp-Glu-Ala-Asp (DEAD). The DEAD box protein family is associated with cellular processes that involve RNA secondary structure alteration such as RNA splicing, ribosome assembly, Initiation of translation, Nuclear and mitochondrial splicing, Spermatogenesis, embryogenesis, and cell growth and division. The RASAL 1 protein is member of GAP1 family that function in suppressing Ras function by inactivating GDP-bound form of Ras which permit the control of cellular proliferation and differentiation. DTX1 function as ubiquitin ligase protein by facilitating ubiquitination and allowing degradation of MEKK1. The ubiquitin ligase activity of DTX1 regulates the Notch Pathway, a signaling pathway that is associated with cell-cell communications that regulates cell-fate determination.
Conservation
The Basic Alignment Search Tool (BLAST)[8] of human CCDC42B protein-to-protein database including Mammalia for closely related species, and excluded Mammalia for distantly related species resulted in several orthologs species with reasonable E-value, and high, medium and low coverage depending on the relatedness of orthologs to human CCDC42B. Higher conservation of CCDC42B gene resulted in several strict orthologs (mammalian) of percentage identity range of 95%-53%: rhesus monkey, whale, pig, cattle, and mouse. Lower conservation of CCDC42B gene in distant homologs (non-mammalian) of percentage identity range of 23%-40%: Drosophila, reptile, amphibians and fish.
Paralogs
CCDC42B gene has only one major paralogs CCDC42'(CCDC42A)
| Name | Species | Species common name | NCBI accession number | Length | Protein identity |
|---|---|---|---|---|---|
| CCDC42B | Homo sapiens | Human | NM_001144872.1 | 308aa | 100% |
| CCDC42A | Homo sapiens | Human | NM_144681.2 | 316aa | 36% |
Orthologs
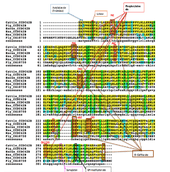
Human CCDC42B gene is found in ~58 orthologs species.[5] CCDC42B higher conservation in many mammalian orthologs species compared to non-mammalian orthologs species. Higher conservation of CCDC42B gene in several strict orthologs (mammalian): chimpanzee, rhesus monkey, dog,cow, mouse, rat and chicken, and identities that range between 95%-69%. Lower conservation of CCDC42B gene in distant homologs (non-mammalian): birds, reptile, amphibians and fish and identities that range between 23%-40%. The figure shows comparison between strict orthologs and distant homologs for conservation of CCDC42B (purple color: matched amino acid residues ; blue: conserved residues ; pink: similar residues ; white: different residues )

| Genus/species | Common name | Class | MYA | Length (AA) | Identity | Accession(RefSeq) | |
| Macaca mulatta | Rhesus monkey | Mammalia | 29 | 308 | 95% | NP_001181192.1 | |
| Orcinus orca | Killer whale | Mammalia | 94.2 | 309 | 81% | XM_004281459.1 | |
| Bos taurus | Cattle | Mammalia | 94.2 | 314 | 79% | NM_001144873.1 | |
| Sus scrofa | Pig | Mammalia | 94.2 | 308 | 79% | XM_005670689.1 | |
| Ceratotherium simum simum | Southern white rhinoceros | Mammalia | 94.2 | 311 | 79% | XM_004430130.1 | |
| Loxodonta africana | African savanna elephant | Mammalia | 98.7 | 303 | 79% | XM_003419288.1 | |
| Trichechus manatus latirostris | Florida manatee | Mammalia | 98.7 | 303 | 78% | XM_004379058.1 | |
| Equus caballus | Horse | Mammalia | 94.2 | 306 | 76% | ||
| Dasypus novemcinctus | Nine-banded armadillo | Mammalia | 104.2 | 310 | 74% | XM_004456157.1 | |
| Microtus ochrogaster | Prairie vole | Mammalia | 92.3 | 308 | 69% | XM_005371872.1 | |
| Ciona intestinalis | Vase tunicate | Ascidiacea | 722.5 | 308 | 40% | XM_002128423.1 | |
| Strongylocentrotus purpuratus | Purple sea urchin | Echinoidea | 742.9 | 312 | 40% | ||
| Xenopus (Silurana) tropicalis | Western clawed frog | Amphibia | 371.2 | 326 | 38.7 | XM_004910626.1 | |
| Crassostrea gigas | Pacific oyster | Bivalvia | 782.7 | 312 | 38.2% | JH816130.1 | |
| Lepisosteus oculatus | Spotted gar | Bony fish | 400.1 | 302 | 38% | XM_006640471.1 | |
| Hydra vulgaris | Fresh-water polyp | Hydrozoa | 855.3 | 305 | 38% | XM_004206385.1 | |
| Chrysemys picta bellii | Western painted turtle | Reptilia | 296 | 321 | 37% | XM_005309857.1 | |
| Anolis carolinesis | Green anole | Reptilia | 296 | 314 | 36% | XM_003217075.1 | |
| Latimeria chalumnae | Coelacanth | bony fish | 414.9 | 312 | 35% | XM_006005425.1 | |
| Amphimedon queenslandica | Sponge | demospongiae | 716.5 | 319 | 33% | XM_003385188.1 | |
| Drosophila melanogaster | Fruit fly | Insecta | 782.7 | 331 | 23% | NP_609955.1 |
Phylogeny
According to Biology Workbench,[9] a phylogenetic tree was constructed showing the divergent of CCDC42B across species.The percent identity vs. the divergent time of orthologs species compared to human sequence is shown below. The figure illustrates the evolutionary history of CCDC42B gene in various species (shown in the orthologs space). The closely related species has higher percent identity, which provides statistical evidence for higher amino acids conservation.Distantly related species to human CCDC42B showed lower percent identity, which supports the few conservation of amino acid residue. The figure highlights the amount of changes occurred in CCDC42B evolution and rate of mutation in the gene.
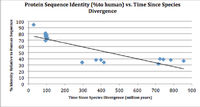
Protein
According to SAPS tool,[9] Human CCDC42B protein is composed of 308 amino acids of 8 exons. The mature form of CCDC42B protein has molecular weight of 35.9 kdal (35,914 Da). The isoelectric point for human CCDC42B is 7.01, in which CCDC42B protein carries no net charge at that particular pH. The N-terminal of the protein sequence is composed of Met (M). The grand average of hydropathicity was predicted to be -0.694 for CCDC42B (Human) and -0.398 for Drosophila melanogaster CG10750, distantly related orthologs. The negative GRAVY confirms that both proteins are soluble and hydrophilic. The theoretical instability index (II) for CCDC42B is predicted to be 63.73 and for CG10750 is 45.20, which indicate that, both proteins are instable in a test tube. The half-life of is predicted to be 30 hours for both CCDC42B and CG10750 in mammalian reticulocytes (in vitro), which correspond to half-life for enzymes responsible for controlling metabolic rate. The above results confirmed that both CCDC42B and CG10750 share similarities in amino acid composition and protein characteristics. Thus, many characteristics of CCDC42B have been conserved across closely and distantly related species.
Primary sequence & variants/isoforms
Human CCDC42B gene contains 9 introns and 8 different mRNA transcripts are produced: 4 alternatively spliced variants and 4 un-spliced variants. Alternative splicing results in encoding 2 very good proteins, 3 good proteins and 3 non-coding proteins.[10]
Domains and motifs
CCDC42B protein of unknown function contains coiled-coil domain of unknown function (DUF4200) that belongs to Eukaryote family and located at range of 34-159 amino acids. The DUF4200 domain has been conserved in Eukaryote. Coiled coil structure consists of two alpha helices wrapped around each other to form a twist. Heptad repeat pattern (abcdefg)n forms the sequence of coiled coil structure, where a and d are hydrophobic, e and g are polar of charged.
| Tool | domains and motifs | Position (AA) |
| 2ZIP [11] | Leucine Zipper domain | 123-154 |
| 2ZIP [11] | coiled-coil | 123-150 & 171-201 |
| PFSCAN[12] | Arginine-rich | 94-139 |
Post-translational modifications
ExPASy Proteomics Tool[13] was primarily used to analyze post-transcriptional modifications of CCDC42B protein. Human CCDC42B N-terminus Acetylation (A2) corresponded in 5 out of 6 orthologs. Drosophila has no Ala, Gly, Ser or Thr at position 1-3, thus N-terminus acetylation is conserved in human CCDC42B. Human CCDC42B protein has conserved SUMOylation site, since lysine (K) at position 285 was conserved in 5 out of 6 orthologs, mostly closely related organisms showed the conservation of lysine. Phosphorylation events occur mostly in CCDC42B, which is suggested to be involved in signaling pathways. Human CCDC42B phosphorylation site of tyrosine at position 8 (Y8) was fully conserved in all 6 orthologs species (the site corresponded with sulfation site). Also other phosphorylation sites in the human CCDC42B protein were conserved in the orthologs (illustrated in the multiple sequence alignment). The same amino acid residues in human CCDC42B protein are subjected to competing phosphorylation and O-linked glycosylation.However, glycosylation sites occur mostly in serine and threonine residues that would be phosphorylated by serine/ threonine kinases. Thus, phosphorylation of the Ser/Thr residues would prevent O-GlcNAc from processing. Human CCDC42B protein has conserved GPI-modification site of Alanine (A) at position 293 that was conserved in 4 out of 6 orthologs.
| Tool | Predicted Modification | Homo sapiens | Mus musculus | Drosophila melanogaster |
| YinOYang[14] | O-β-GlcNAc | T60, T240, S308 | T302, T304, T306 | S30, S116, T155, S238, S241 |
| NetPhos[15] | phosphorylation | S18, S80, T227, T277, Y8 | S14, S58, S170, S188, S198, S238, S240, T4, T25, T59, T119, T167, T269 | S19, S45, S116, S120, S141, S178, S201, S238, S241, S261, S290, S293, S308, S319, T7, T125, T132, Y239 |
| Sulfinator[16] | sulfation | (none) | (none) | Y61 |
| SulfoSite[17] | sulfation | Y8 | Y56 | Y61,Y294 |
| SumoPlot[18] | sumoylation | K289 | K178, K287, K202, K53, K38, K39, K153 | K9, K251, K232, K39, K328, K99 |
| Terminator[19] | N-terminus | A2 | A2 | P2 |
Secondary structure
CCDC42B protein form a secondary structure based upon alpha-helices. The structure of CCDC42B is predicted to contain several alpha-helices, and other random coils. Hairpin loop structures were detected at the 5'UTR and 3'UTR region of CCDC42B. Also, leucine zipper domain was found overlapping with coiled-coil domain. The attached image shows comparison between human CCDC42B and 5 other orthologs species which supports that human CCDC42B is primarily composed of alpha helices for its secondary structure.
3° and 4° structure
According to CBLAST,[20] the CCDC42B protein sequence was aligned with 2I1K_A (Chain A, Moesin From Spodoptera Frugiperda Reveals The Coiled-Coil Domain At 3.0 Angstrom Resolution), and an E-value of 1.00e-03 was obtained. The aligned sequences from 164-243 AA for CCDC42B, and 302-381 AA for 2I1K_A resulted in 22% identity between both sequences in 80 amino acid residues.The structure shows only the aligned sequence of CCDC42B with 2I1K_A. Predicted structure (blue: not similar residues, red: conserved residues, gray: not aligned CCDC42B residues with 2I1K_A).
Expression
Human Protein atlas [21] resulted in CCDC42B expression in normal human tissue. The expression level of CCDC42B gene in human normal tissues was detected at high to moderate level in 17 out of 78 tissues analyzed using Expressed Sequence Tag (EST) technique. CCDC42B gene has a narrowed expression in tissues. The gene has higher expression in respiratory epithelia and fallopian tube; Moderate expression in intestine and liver; and low to none expression in other normal tissues. Moreover, Microarray and Immunohistochemistry (IHC) expression detected presence of low level of CCDC42B mRNA expression in: salivary gland, stomach, skin, bone marrow, and lung. Coiled coil domain containing 42B is involved in cancer; CCDC42B gene is expressed in low to moderate level in tumor cell.
Promoter and Transcription Binding Factors

According to Genomatix,[22] the Promoter region contains 859 base pairs and it is located on the positive strand of chromosome 12 from region 113,586,906 to 113,587,764 upstream of CCDC42B gene. The promoter region was predicted to contain sites for transcription binding factors that regulate expression of CCDC42B. The Attached image illustrate important transcription binding factors in the promoter region for human CCDC42B .
Expression
CCDC42B gene has a narrowed expression in tissues. The gene has higher expression in respiratory epithelia and fallopian tube; Moderate expression in intestine and liver; low to none expression in other normal tissues. Coiled coil domain containing 42B is involved in some types of cancer. CCDC42B gene is expressed in low to moderate level in tumor cell.[21][23][24]
Function / Biochemistry
According to year 2014, CCDC42B gene/protein has unknown function in homo sapiens. However, Human CCDC42B is predicted to be involved in flagella assembly and motility.
Interacting Proteins
According to STRING,[25] MINT,[26] and IntAct,[27] Human CCDC42B did not show any direct interaction with other proteins. Searching GeneMania,[28] other interactions have been identified by co-expression with other proteins as seen in the figure. CCDC42B was found to co-express with other coiled-coil domains containing proteins (CCDC78 and CCDC153). Since Human CCDC42B is expressed in low level in testis, it is predicted that human to interact with SPATC1 (Spermatogenesis and centriole associated 1).
Clinical Significance
Disease Association
Human CCDC42B is located at chromosome 12 (12q24.13), which is linked to skeletal deformities, hypochondrogenesis, achondrogenesis, and kniest dysplasia. According to OMIM[29] search chromosome 12 (12q24.1) is linked Noonan syndrome 1 that is caused by heterozygote mutation in PTPN11 gene product, SH-PTP2, and primarily causing facial developmental defects and heart defects.
Mutations
Two SNPs (Y8, Q280) are highly conserved in many orthologs species. Thus, these residues can change function of protein leading to possible disease not only in human.
| SNP | Chromosome (12) | Position | Region of gene | Type | Allele change | Residue change | |
|---|---|---|---|---|---|---|---|
| Rs61748300 | 113587667 | 2 | CDS region | Missense (Non-synonymous) | GCG→GTG | Ala (A)→Val(V) | |
| Rs373892417 | 113587685 | 8 | CDS region | Missense (Non-synonymous) | TAT→TGT | Tyr (Y)→Cys (C) | |
| Rs61738699 | 113589799 | 45 | CDS region | Missense(Non-synonymous) | GCA→ACA | Ala (A)→Thr (T) | |
| Rs377463846 | 113590594 | 57 | CDS region | Missense(Non-synonymous) | CGC→ TGC | Arg(R) → Cys (C) | |
| Rs34765757 | 113591023 | 94 | CDS region | Frame shift | CGG→ G | Arg (R) → Gly (G) | |
| Rs34276842 | 113591036 | 98 | CDS region | Frame shift | GCG→ CG Ala | (A)→ Arg (R) | |
| Rs370323183 | 113591110 | 122 | CDS region | Missense(Non-synonymous) | CAG→CGG | Gln (Q) → Arg (R) | |
| Rs34078446 | 113591152 | 138 | CDS region | Frame shift | AAG→A | Lys (K)→ Ser (S) | |
| Rs200344876 | 113592306 | 187 | CDS region | Frame shift | →GGA | Glu (E) → Gly (G) | |
| Rs377537662 | 113593122 | 250 | CDS region | Missense(Non-synonymous) | CGC→TGC | Arg (R)→Cys (C) | |
| RS144548708 | 113593212 | 280 | CDS region | Missense(Non-synonymous) | CAG→GAG | Gln (Q)→Glu (E) |
Conceptual Translation
Major predicted domains, post-transcriptional modification sites, and structural form are shown in the conceptual translation


References
- GRCh38: Ensembl release 89: ENSG00000186710 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000094282 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "CCDC42B coiled-coil domain containing 42B [ Homo sapiens (human) ]". National Center for Biotechnology Information (NCBI). Retrieved 17 March 2014.
- "Coiled-Coil Domain Containing 42B". GeneCards. Retrieved 18 March 2014.
- "Homo sapiens gene CCDC42B, encoding coiled-coil domain containing 42B". National Center for Biotechnology Information. Retrieved 18 March 2014.
- "BLAST". National Center for Biotechnology Information Search database (NCBI). Archived from the original on 4 May 2014. Retrieved 6 May 2014.
- "ClustLaw". Biology WorkBench. Retrieved 6 May 2014.
- "Homo sapiens gene CCDC42B, encoding coiled-coil domain containing 42B". AceView (NCBI). Retrieved 6 May 2014.
- Erich Bornberg-Bauer; Eric Rivals; Martin Vingron. "2ZIP". Computational Approaches to Identify Leucine Zippers. Retrieved 13 April 2014.
- "Sequence Search Against a Set of Profiles (PROSITE and PFAM)". Biology WorkBench.
- . ExPASy: SIB Bioinformatics Resource Portal http://www.expasy.org/proteomics/families__patterns_and_profiles. Retrieved 6 May 2014. Missing or empty
|title=(help) - "YinOYang". Retrieved 12 Apr 2014.
- "NetPhos". Retrieved 12 Apr 2014.
- "Sulfinator". Retrieved 12 Apr 2014.
- "SulfoSite". Archived from the original on 2008-07-24. Retrieved 12 Apr 2014.
- "SumoPlot". Archived from the original on 20 April 2009. Retrieved 12 Apr 2014.
- "Terminator". Archived from the original on 2008-04-16. Retrieved 12 Apr 2014.
- "CCDC42B". Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, Zhang N, Bryant SH (2007), "MMDB: annotating protein sequences with Entrez's 3D-structure database", Nucleic Acids Res.35(D)205-10. Retrieved 6 May 2014.
- "CCDC42B". Human Protein Atlas. Retrieved 6 May 2014.
- . © Genomatix Software GmbH 2014 http://www.genomatix.de/. Retrieved 6 May 2014. Missing or empty
|title=(help) - "Expression of CFAP73 in cancer". The Human Protein Atlas.
- "Cilia- and flagella-associated protein 73 Expression". Nextprot Beta.
- "STRING - Known and Predicted Protein-Protein Interactions". STRING. Retrieved 6 May 2014.
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, et al. (January 2012). "MINT, the molecular interaction database: 2012 update". Nucleic Acids Research. 40 (Database issue): D857–61. doi:10.1093/nar/gkr930. PMC 3244991. PMID 22096227.
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, et al. (January 2014). "The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases". Nucleic Acids Research. 42 (Database issue): D358–63. doi:10.1093/nar/gkt1115. PMC 3965093. PMID 24234451.
- "CCDC42B". genemania. Retrieved 10 May 2014.
- "OMIM". National Center for Biotechnology Information (NCBI). Retrieved 6 May 2014.