Cation-dependent mannose-6-phosphate receptor
In the fields of biochemistry and cell biology, the cation-dependent mannose-6-phosphate receptor (CD-MPR) also known as the 46 kDa mannose 6-phosphate receptor is a protein that in humans is encoded by the M6PR gene.[5][6]
The CD-MPR is one of two transmembrane proteins that bind mannose-6-phosphate (M6P) tags on acid hydrolase precursors in the Golgi apparatus that are destined for transport to the lysosome. Homologues of CD-MPR are found in all eukaryotes.
Structure
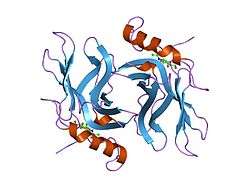
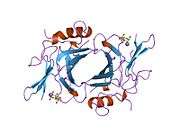
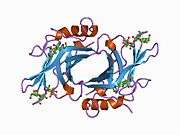
The CD-MPR is a type I transmembrane protein (that is, it has a single transmembrane domain with its C-termini on the cytoplasmic side of lipid membranes) with a relatively short cytoplasmic tail.[7] The extracytoplasmic/lumenal M6P binding-domain consists of 157 amino acid residues. The CD-MPR is approximately 46 kDa in size and it both exists and functions as a dimer.
The cell surface receptor for insulin-like growth factor 2 also functions as a cation-independent mannose 6-phosphate receptor.[7] It consists of fifteen repeats homologous to the 157-residue CD-M6PR domain, two of which are responsible for binding to M6P.
Function
Both CD-MPRs and CI-MPRs are lectins that bind their M6P-tagged cargo in the lumen of the Golgi apparatus. The CD-MPR shows greatly enhanced binding to M6P in the presence of divalent cations, such as manganese.[7] The MPRs (bound to their cargo) are recognized by the GGA family of clathrin adaptor proteins and accumulate in forming clathrin-coated vesicles.[8] They are trafficked to the early endosome where, in the relatively low pH environment of the endosome, the MPRs release their cargo. The MPRs are recycled back to the Golgi, again by way of interaction with GGAs and vesicles. The cargo proteins are then trafficked to the lysosome via the late endosome independently of the MPRs.
See also
References
- GRCh38: Ensembl release 89: ENSG00000003056 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000007458 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Dahms NM, Lobel P, Breitmeyer J, Chirgwin JM, Kornfeld S (July 1987). "46 kd mannose 6-phosphate receptor: cloning, expression, and homology to the 215 kd mannose 6-phosphate receptor". Cell. 50 (2): 181–92. doi:10.1016/0092-8674(87)90214-5. PMID 2954652.
- Pohlmann R, Nagel G, Schmidt B, Stein M, Lorkowski G, Krentler C, Cully J, Meyer HE, Grzeschik KH, Mersmann G (August 1987). "Cloning of a cDNA encoding the human cation-dependent mannose 6-phosphate-specific receptor". Proc. Natl. Acad. Sci. U.S.A. 84 (16): 5575–9. doi:10.1073/pnas.84.16.5575. PMC 298905. PMID 2441386.
- Ghosh P, Dahms NM, Kornfeld S (March 2003). "Mannose 6-phosphate receptors: new twists in the tale". Nat. Rev. Mol. Cell Biol. 4 (3): 202–12. doi:10.1038/nrm1050. PMID 12612639.
- Ghosh P, Kornfeld S (July 2004). "The GGA proteins: key players in protein sorting at the trans-Golgi network". Eur. J. Cell Biol. 83 (6): 257–62. doi:10.1078/0171-9335-00374. PMID 15511083.
Further reading
- Yang EB, Qin LL, Zhao YN, Zhang K, Chow P (2003). "Genetic changes and expression of the mannose 6-phosphate/insulin-like growth factor II receptor gene in human hepatitis B virus-associated hepatocellular carcinoma". Int. J. Mol. Med. 11 (6): 773–8. doi:10.3892/ijmm.11.6.773. PMID 12736721.
- Shin BK, Wang H, Yim AM, Le Naour F, Brichory F, Jang JH, Zhao R, Puravs E, Tra J, Michael CW, Misek DE, Hanash SM (2003). "Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function". J. Biol. Chem. 278 (9): 7607–16. doi:10.1074/jbc.M210455200. PMID 12493773.
- Lemansky P, Fester I, Smolenova E, Uhländer C, Hasilik A (2007). "The cation-independent mannose 6-phosphate receptor is involved in lysosomal delivery of serglycin". J. Leukoc. Biol. 81 (4): 1149–58. doi:10.1189/jlb.0806520. PMID 17210618.
- Doray B, Bruns K, Ghosh P, Kornfeld S (2002). "Interaction of the cation-dependent mannose 6-phosphate receptor with GGA proteins". J. Biol. Chem. 277 (21): 18477–82. doi:10.1074/jbc.M201879200. PMID 11886874.
- Schimanski LM, Drakesmith H, Sweetland E, Bastin J, Rezgui D, Edelmann M, Kessler B, Merryweather-Clarke AT, Robson KJ, Townsend AR (2009). "In vitro binding of HFE to the cation-independent mannose-6 phosphate receptor". Blood Cells Mol. Dis. 43 (2): 180–93. doi:10.1016/j.bcmd.2009.03.010. PMID 19487139.
- Rodriguez-Gabin AG, Yin X, Si Q, Larocca JN (2009). "Transport of mannose-6-phosphate receptors from the trans Golgi network to endosomes requires Rab31". Exp. Cell Res. 315 (13): 2215–30. doi:10.1016/j.yexcr.2009.03.020. PMC 2753287. PMID 19345684.
- Seaman MN (2004). "Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer". J. Cell Biol. 165 (1): 111–22. doi:10.1083/jcb.200312034. PMC 2172078. PMID 15078902.
- Kölsch H, Ptok U, Majores M, Schmitz S, Rao ML, Maier W, Heun R (2004). "Putative association of polymorphism in the mannose 6-phosphate receptor gene with major depression and Alzheimer's disease". Psychiatr. Genet. 14 (2): 97–100. doi:10.1097/01.ypg.0000129204.58574.c2. PMID 15167696.
- Xaplanteri P, Lagoumintzis G, Dimitracopoulos G, Paliogianni F (2009). "Synergistic regulation of Pseudomonas aeruginosa-induced cytokine production in human monocytes by mannose receptor and TLR2". Eur. J. Immunol. 39 (3): 730–40. doi:10.1002/eji.200838872. PMID 19197942.
- Hille-Rehfeld A (1995). "Mannose 6-phosphate receptors in sorting and transport of lysosomal enzymes". Biochim. Biophys. Acta. 1241 (2): 177–94. doi:10.1016/0304-4157(95)00004-b. PMID 7640295.
- Pérez-Victoria FJ, Mardones GA, Bonifacino JS (2008). "Requirement of the Human GARP Complex for Mannose 6-phosphate-receptor-dependent Sorting of Cathepsin D to Lysosomes". Mol. Biol. Cell. 19 (6): 2350–62. doi:10.1091/mbc.E07-11-1189. PMC 2397299. PMID 18367545.
- Munier-Lehmann H, Mauxion F, Hoflack B (1996). "Function of the two mannose 6-phosphate receptors in lysosomal enzyme transport". Biochem. Soc. Trans. 24 (1): 133–6. doi:10.1042/bst0240133. PMID 8674621.
- Mari M, Bujny MV, Zeuschner D, Geerts WJ, Griffith J, Petersen CM, Cullen PJ, Klumperman J, Geuze HJ (2008). "SNX1 defines an early endosomal recycling exit for sortilin and mannose 6-phosphate receptors". Traffic. 9 (3): 380–93. doi:10.1111/j.1600-0854.2007.00686.x. PMID 18088323.
- Op den Brouw ML, Binda RS, Geijtenbeek TB, Janssen HL, Woltman AM (2009). "The mannose receptor acts as hepatitis B virus surface antigen receptor mediating interaction with intrahepatic dendritic cells". Virology. 393 (1): 84–90. doi:10.1016/j.virol.2009.07.015. PMID 19683778.
- Nair P, Schaub BE, Rohrer J (2003). "Characterization of the endosomal sorting signal of the cation-dependent mannose 6-phosphate receptor". J. Biol. Chem. 278 (27): 24753–8. doi:10.1074/jbc.M300174200. PMID 12697764.
- Kato N, Miyata T, Tabara Y, Katsuya T, Yanai K, Hanada H, Kamide K, Nakura J, Kohara K, Takeuchi F, Mano H, Yasunami M, Kimura A, Kita Y, Ueshima H, Nakayama T, Soma M, Hata A, Fujioka A, Kawano Y, Nakao K, Sekine A, Yoshida T, Nakamura Y, Saruta T, Ogihara T, Sugano S, Miki T, Tomoike H (2008). "High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project". Hum. Mol. Genet. 17 (4): 617–27. doi:10.1093/hmg/ddm335. PMID 18003638.
- Chen JJ, Zhu Z, Gershon AA, Gershon MD (2004). "Mannose 6-phosphate receptor dependence of varicella zoster virus infection in vitro and in the epidermis during varicella and zoster". Cell. 119 (7): 915–26. doi:10.1016/j.cell.2004.11.007. PMID 15620351.
External links
- cation-dependent+mannose-6-phosphate+receptor at the US National Library of Medicine Medical Subject Headings (MeSH)