Bovine foamy virus
Bovine foamy virus (BFV) is a ss(+)RNA retrovirus that belongs to the genus spumaviridae. Spumaviruses are differ from the other six members of family retroviridae, both structurally and in pathogenic nature. Supma viruses derive their name from spuma the latin for "foam". The 'foam' part of 'foamy virus' comes from syncytium formation and the rapid vacuolization of infected cells which creates a 'foamy' appearance.[1][2]
| Bovine foamy virus | |
|---|---|
| Virus classification | |
| Group: | Group VI (ssRNA-RT) |
| Order: | |
| Family: | |
| Subfamily: | Spumaretrovirinae |
| Genus: | |
| Species: | Bovine foamy virus |
| Serotype | |
|
Bovine foamy virus | |
Discovery
The first foamy viral agent was isolated from liver structures of the Rhesus macaque in 1955.[3][4] Bovine foamy virus was isolated in 1969 from cattle with lymphosarcoma.[5] At the time of its discovery little was known about the effects of the virus or whether it played any role whatsoever in the development of lymphosarcoma in the cattle. Continued research has shown that the infection prevalence of BFV is 40-85% worldwide but lacks any significant pathogenic nature.[6][5]
Structure/Morphology
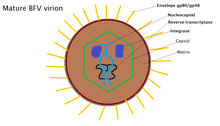
Bovine foamy virus is an enveloped, spherical virus. The virus particles are approximately 80-100 nm in diameter and contain single-strand, positive-sense RNA: ss(+)RNA. 20% of released virions contain double stranded DNA as a product of late stage reverse transcription. The outer membrane of the virus is covered with a variety of glycoproteins which allow it to interact with its surroundings. Just beyond the envelope is a buffer between the capsid and envelope, known as the protein matrix. The matrix is responsible for maintaining membrane shape as well as participating in the budding process. Inside the matrix lies the capsid, a protein shell that contains viral intigrase, reverse-transcriptase and the nucleocapsid, inside of which lives the viruses' genetic material.[7] Due to the viruses' tendency to bud into the endoplasmic reticulum, the virions tend to display immature protein characteristics around the core as well as unique glycoptorien spikes on the surface of the membrane. BFV particles also possess electron-lucent membranes which are easily cross-able by electrons.[1]
Viral genome
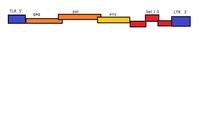
BFV has a monoparatie genome, meaning that its entire genome is stored in a single ssRNA(+)/dsDNA molecule. This molecule is linear dimeric genome contain approximate 12.3kb of information. The molecule possesses an 5'-cap and 3'poly-A tail. Each of these ends possesses a 600nt long sequence of terminals repeats (LTR). The U3, R ad U5 regions are contained within these LTRs. The 5' end possesses a primer binding site while the 3' end possesses a polypurine tract.[7]
Viral entry
Attachment
The process by which foamy viruses enter a cell lacks certain levels of characterization, however it is known that the virus particle first attaches to its host using the SU glycoprotein present on its envelope.[8] The binding receptor for foamy viruses is not entirely know, however it is known that the virus is capable of infecting a board spectrum of cells, meaning the binding receptor must be quite prevalent in most forms of cells.[9] Research has shown that the most likely candidate for a common receptor is heparan sulfate (HS), an extremely common glycosaminoglycan (GAG) present on the ECM of many cells.[9]
Fusion
Once internalized, the virus particle must fuse with the internal vessical and allow its genetic material to escape into the cytoplasm. Viral fusion is mediated through the TM glycoprotein. The fusion process is imitated when the TM protein is cleaved by the changing levels of pH which in turns creates a conformation change in the protein, fusing the membrane of the virus with the internal vessical.[9]
Replication cycle
Cycle overview
Once the viral particle has delivered its genetic material safely into the cytoplasm of its host, one of two things will occur. If the genetic payload of the virion was double stranded DNA, the viral DNA will directly integrate into the host's genome via the viral enzyme integrase.[1][7] If the payload was ssRNA the RNA is first copied into sdDNA via reverse transcriptase and then integrated randomly into the host genome by integrase. The host cell then performs its natural replication functions, however while doing so it transcribes viral RNA which is then translated into precursor polyproteins.[7] At this point in the cycle, if any viral RNA is remaining, it will be transcribed into dsDNA, however it is too late to be integrated into the host-cell genome. This dsDNA is then randomly packaged into newly formed virions along with the ssRNA. From this point the viral particles begin to bud. Most bud into the endoplasmic reticulum which are then packaged and distributed to other cells. Some particles manage to escape through the membrane but this number is usually quite low.[7][10][11]
Unique features of the replication cycle
Being a spumavirus, bovine foamy virus has a unique life cycle, even when compared to other retroviruses. One of these differences is the mechanism used in the budding process. Rather than budding through the plasma membrane like a more traditional retrovirus, BFV and other foamy viruses bud using the endoplasmic reticulum. Another facet of BFV that makes it unique among other retroviruses is how late in the replication cycle reverse-transcription takes place. This results in some of virions containing DNA rather than typical RNA. This also results in some level of integration into the host cell DNA in uncommon ways.[10] For this reason, scientists have likened foamy viruses to viruses of the family Hepadnaviridae. Foamy viruses also contain structural genes that are distinct to their particular genus. The Gag protein is not always correctly cleaved into the mature form of the viral protein, as seen in other members of retrovirdae, which leads to the overall immature morphology of FV.[1] The Env budding associated protein found in the transmembrane domain of the viron contains an endoplasmic reticulum retention signal which contributes to its ability to bud into the ER.[1][11]
Host interactions
The most obvious result of interactions between the virus and its host is the telltale formation of large numbers of vacuoles throughout the cytoplasm. This is referred to as vacuolization, and it is what gives the spumaviruses their name. Most infected cells also begin rapid syncytium formation.[1] Thought this effect in seen quite frequently, its cause is still yet to be determined. In some more rare instances, cell death has been recorded.[2]
Associated diseases
To date, no known pathogen is associated with bovine foamy virus.
Tropism
Bovine foamy virus is known to be capable of infecting a wide range of cells.[2] These include fibroblasts, epithelial cells and neural cells. Using techniques such as PCR, bovine foamy virus can be identified in most tissues of infected animals.[1]
References
- Meiering, Christopher D.; Maxine L. Linial (January 2001). "Historical Perspective of Foamy Virus Epidemiology and Infection". Clinical Microbiology Reviews. 14 (1): 165–176. doi:10.1128/CMR.14.1.165-176.2001. ISSN 0893-8512. PMC 88968. PMID 11148008.
- Linial, Maxine L. (1999-03-01). "Foamy Viruses Are Unconventional Retroviruses". Journal of Virology. 73 (3): 1747–1755. doi:10.1128/JVI.73.3.1747-1755.1999. ISSN 0022-538X. PMC 104413. PMID 9971751.
- Enders, J. F.; Peebles, T. C. (June 1954). "Propagation in tissue cultures of cytopathogenic agents from patients with measles". Proceedings of the Society for Experimental Biology and Medicine (New York, N.Y.). 86 (2): 277–286. doi:10.3181/00379727-86-21073. ISSN 0037-9727. PMID 13177653.
- Rustigian, R.; Johnston, P.; Reihart, H. (January 1955). "Infection of monkey kidney tissue cultures with virus-like agents". Proceedings of the Society for Experimental Biology and Medicine (New York, N.Y.). 88 (1): 8–16. doi:10.3181/00379727-88-21478. ISSN 0037-9727. PMID 14357329.
- Malmquist, W. A.; Van der Maaten, M. J.; Boothe, A. D. (January 1969). "Isolation, immunodiffusion, immunofluorescence, and electron microscopy of a syncytial virus of lymphosarcomatous and apparently normal cattle". Cancer Research. 29 (1): 188–200. ISSN 0008-5472. PMID 4974302.
- Johnson, R. H.; de la Rosa, J.; Abher, I.; Kertayadnya, I. G.; Entwistle, K. W.; Fordyce, G.; Holroyd, R. G. (January 1988). "Epidemiological studies of bovine spumavirus". Veterinary Microbiology. 16 (1): 25–33. doi:10.1016/0378-1135(88)90124-1. ISSN 0378-1135. PMID 2833003.
- SIB Swiss Insitiute of Bioinformatics. "Spuma Virus".
- taxonomy. "Taxonomy Browser". www.ncbi.nlm.nih.gov. Retrieved 2017-11-02.
- Plochmann, Kathrin; Horn, Anne; Gschmack, Eva; Armbruster, Nicole; Krieg, Jennifer; Wiktorowicz, Tatiana; Weber, Conrad; Stirnnagel, Kristin; Lindemann, Dirk (September 2012). "Heparan Sulfate Is an Attachment Factor for Foamy Virus Entry". Journal of Virology. 86 (18): 10028–10035. doi:10.1128/JVI.00051-12. ISSN 0022-538X. PMC 3446549. PMID 22787203.
- Moebes, A.; Enssle, J.; Bieniasz, P. D.; Heinkelein, M.; Lindemann, D.; Bock, M.; McClure, M. O.; Rethwilm, A. (1997-10-01). "Human foamy virus reverse transcription that occurs late in the viral replication cycle". Journal of Virology. 71 (10): 7305–7311. ISSN 0022-538X. PMC 192074. PMID 9311807.
- Goepfert, P. A.; Wang, G.; Mulligan, M. J. (1995-08-25). "Identification of an ER retrieval signal in a retroviral glycoprotein". Cell. 82 (4): 543–544. doi:10.1016/0092-8674(95)90026-8. ISSN 0092-8674. PMC 7172326. PMID 7664333.