Beta-propeller phytase
β-propeller phytases (BPPs) are a group of enzymes (i.e. protein superfamily) with a round beta-propeller structure. BPPs are phytases, which means that they are able to remove (hydrolyze) phosphate groups from phytic acid and its phytate salts.[2] Hydrolysis happens stepwise and usually ends in myo-inositol triphosphate product which has three phosphate groups still bound to it.[3] The actual substrate of BPPs is calcium phytate[4] and in order to hydrolyze it, BPPs must have Ca2+ ions bound to themselves. BPPs are the most widely found phytase superfamily in the environment and they are thought to have a major role in phytate-phosphorus cycling in soil and water.[5] As their alternative name alkaline phytase suggests, BPPs work best in basic (or neutral) environment. Their pH optima is 6–9,[2] which is unique among the phytases.[5]

Potential uses
As of April 2018, BPPs are not used commercially, but they may have potential for such use. Histidine acid phytases (HAPs) are the only group of phytases which are used in animal feed at the moment.
Animal feed
Recombinant phytases are added commonly in agriculture to animal feed of monogastric animals to enhance the feed's nutrient bioavailability.[6] These nutrients include phosphorus which is bound to phytates in the form of their phosphate groups. In contrast to ruminants like cattle, gut bacteria of monogastric animals like pigs and chickens can't properly hydrolyze these groups free so that the digestive system of the animal can use the phosphorus. Unabsorbed phosphorus is thus wasted and may end up into the environment in animal manure via agricultural runoff and cause eutrophication. Phytic acid can also work as an antinutrient: it can chelate calcium from feed and decrease its bioavailability up to 60–70% of the feed's total calcium content. Phytase addition improves calcium availability and can also improve the bioavailability of iron and zinc. It might also increase the availability of copper and manganese. Amino acid bioavailability is not enhanced significantly.[7]
In comparison to histidine acid phytases (HAPs), which are often unstable in temperatures higher than 65 °C, BPPs can naturally withstand high temperatures of 80–85 °C. Such temperatures are commonly used in pelleting of animal feed during its manufacture. Unlike HAPs, BPPs have a neutral or alkaline pH optima, which makes it possible to use them in neutral or alkaline environments. This expands potential applications for phytases.[2] BPPs could be used in aquatic animal feed because many of these animals like fishes and shrimps have a neutral or alkaline gastrointestinal tract.[8] BPPs are also phytate specific unlike HAPs,[5] which hydrolyze also other phosphate containing molecules like ADP, GTP and NADH.[9] However, BPPs are catalytically more than 2–60 times slower than current the HAPs. HAPs have a specific catalytic activity of 100–3000 U mg−1. BPPs usually have a specific catalytic activity of less than 50 U mg−1.[2] Because of such low activity practical use of BPPs requires much more research.[5]
Structure
As of April 2018, 7 BPP crystal structures were known: 3AMR, 3AMS, 1H6L, 1POO, 2POO, 1CVM and 1QLG.
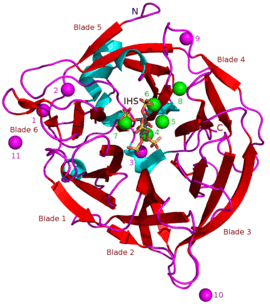
Masses of known BPPs are approximately 35–68 kDa.[5] Their donut-shaped β-propeller structure consists of 6 antiparallel beta sheet structures or "blades". One of these blades has 5 β-sheets (blade number 5 in the picture in the beginning of this article) and the rest have 4 β-sheets. There are hydrophobic interactions between these blades which are thought to keep the propeller structure together. These blades form a tunnel-like hole through the enzyme. This tunnel binds some water molecules. In front of the tunnel is the enzyme's active site which is positively charged in total due to Ca2+ ions it binds and certain positive amino acid residues. This site binds negatively charged calcium phytate and hydrolyzes phosphates from it.[2]
Motifs

A 2008 study by Huang et al. compared 66 BPP peptide sequences and found that sequence motifs DA[A/T/E]DDPA[I/L/V]W and NN[V/I]D[I/L/V]R[Y/D/Q] were conserved in all of the studied BPPs. R[Y/D/Q] for example signifies that R and Y, D or Q were found in the sequence, i.e. RY or RD or RQ.[10] 2014 study by Kumar et al. compared 44 BPPs and found 10 motifs. Two of these, DDPAIW[VI][HN]PK[DN]P[ESA]KS and NN[F/V]D[I/V/L], were found in all of the studied BPPs. These were noted to be similar to the ones found in the 2008 study by Huang et al.[5]
Calcium dependence and inhibitors
BPPs are calcium dependent metalloproteins. Their active site has a number of calcium cations (Ca2+) bound to it via negatively charged amino acid carboxylate groups. Positive calcium ions are needed to make binding of the negative phytate electrically favorable. Binding happens via phytate's negatively charged phosphate groups along with certain positive amino acid residues of the BPP which bind directly to the phytate.[1] Ca2+ concentrations also have an effect on BPP pH optima and thermostability: e.g. with Bacillus sp. KHU-10 BPP the activity is highest with 10 mM of added CaCl2 at 60 °C and pH 6–9.5. Without added CaCl2 the pH highest activity is at 40 °C and pH 6.5–8.5.[5]
Removal of Ca2+ leads to the loss of catalytic activity which is why Ca2+ chelating EDTA inhibits BPPs. Certain point mutations of the calcium binding amino acids also stops enzyme function. Divalent ions like Cd2+, Mn2+, Cu2+, Ba2+, Hg2+, Zn2+, Co2+ and Fe2+ inhibit BPPs by replacing Ca2+ within the enzyme.[5] This is probably due to the fact that these cations are too small in comparison to Ca2+ ions, which have a Van der Waals radius (WDV) of 0.99 Å. Co2+ has VDW of 0.74 Å for example and is thus probably too small to carry out the same tasks that Ca2+ can. However, Sr2+ ions can replace Ca2+ at least in certain cases without total loss of catalytic function. Sr2+ VDW radius is 1.12 Å, and is similar to that of Ca2+.[4] Similar compatibility between different ions can be seen in some other enzymes, too.[11]
High Ca2+ concentration can enhance BPP catalysis rate up to a limit. When Ca2+ concentration surpasses this limit, extra Ca2+ ions begin to work as competitive inhibitors. High concentrations of free phytate which not bound to Ca2+ also inhibits BPPs. This happens possibly via free phytate mediated chelation of the Ca2+ bound to BPPs.[4]
Other types of inhibitors include oxyanionic (oxygen binding) molybdate, tungstate and vanadate. It has been suggested that inhibition with these oxyanions happens because they form trigonal bipyramidal complexes within the active site of the enzyme which are similar to the transition state of the phytate's phosphate group during its hydrolysis. Orthophosphate which is released from phytic acid works as a competitive inhibitor of BPPs.[9] Phytic acid analogue myo-inositol-hexasulphate (IHS) inhibits BPPs and this has been used as an aid in BPP structural studies due to its similarity with phytic acid (see 3AMR).[2]
Hydrolysis mechanism
In a hydrolysis mechanism suggested in 2001 by Shin et al. the Ca2+ ions bound to the BPP are divided to hydrolysis and affinity site ions. In the hydrolysis site, Ca2+ help the phosphate about to be removed bind to the active site. They also activate a water molecule that participates in the hydrolysis by turning it in to an OH− ion and stabilize the transition state during catalysis. Affinity area ions increase phytate affinity to the active site and keep the phytate still during hydrolysis from its other phosphate groups. Hydrolysis is repeated stepwise until a myo-inositol product with three phosphates is obtained.[3] Other studies support product with three phosphates,[12][13][14][15] but more phosphates can be removed under extreme conditions such as high BPP concentration and lengthened incubation time.[13]
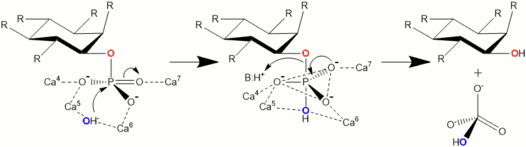
Actual hydrolysis suggested by Shin et al. happens in two steps. Second one is slower and limits the total reaction speed. In the first step carbonyl group withdraws electrons from phosphorus of the phosphate making it electron poor, i.e. leaving it with a positive charge. Simultaneously OH− donates an electron pair to the nominally positive phosphorus in phosphate. A trigonal bipyramidal intermediate state is formed. In the second step phosphoester bond is cleaved when an acidic amino acid residue (BH+) donates a proton to the oxygen in the bond. Phosphate group is thus cleaved off.[3]
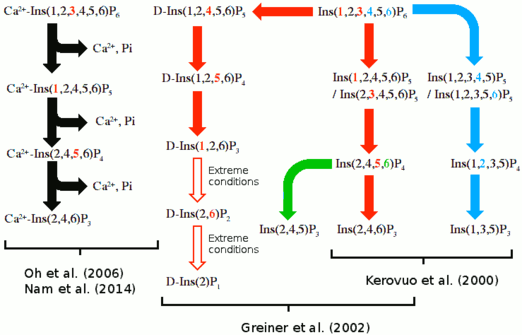
Hydrolysis routes
Multiple suggested hydrolysis routes exists, but it is uncertain which one is correct or if multiple routes exist. These hydrolysis routes are summarized below.
References
- Zeng YF, Ko TP, Lai HL, Cheng YS, Wu TH, Ma Y, Chen CC, Yang CS, Cheng KJ, Huang CH, Guo RT, Liu JR (June 2011). "Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate". Journal of Molecular Biology. 409 (2): 214–24. doi:10.1016/j.jmb.2011.03.063. PMID 21463636.
- Chen C, Cheng K, Ko T, Guo R (2015-04-01). "Current Progresses in Phytase Research: Three-Dimensional Structure and Protein Engineering". ChemBioEng Reviews. 2 (2): 76–86. doi:10.1002/cben.201400026.
- Shin S, Ha NC, Oh BC, Oh TK, Oh BH (September 2001). "Enzyme mechanism and catalytic property of beta propeller phytase". Structure. 9 (9): 851–8. doi:10.1016/S0969-2126(01)00637-2. PMID 11566134.
- Ha NC, Oh BC, Shin S, Kim HJ, Oh TK, Kim YO, Choi KY, Oh BH (February 2000). "Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states". Nature Structural Biology. 7 (2): 147–53. doi:10.1038/72421. PMID 10655618.
- Kumar V, Yadav AN, Verma P, Sangwan P, Saxena A, Kumar K, Singh B (May 2017). "β-Propeller phytases: Diversity, catalytic attributes, current developments and potential biotechnological applications". International Journal of Biological Macromolecules. 98: 595–609. doi:10.1016/j.ijbiomac.2017.01.134. PMID 28174082.
- Gifre L, Arís A, Bach À, Garcia-Fruitós E (March 2017). "Trends in recombinant protein use in animal production". Microbial Cell Factories. 16 (1): 40. doi:10.1186/s12934-017-0654-4. PMC 5336677. PMID 28259156.
- Lei XG, Weaver JD, Mullaney E, Ullah AH, Azain MJ (January 2013). "Phytase, a new life for an "old" enzyme". Annual Review of Animal Biosciences. 1 (1): 283–309. doi:10.1146/annurev-animal-031412-103717. PMID 25387021.
- Viader-Salvadó JM, Gallegos-López JA, Carreón-Treviño JG, Castillo-Galván M, Rojo-Domínguez A, Guerrero-Olazarán M (October 2010). "Design of thermostable beta-propeller phytases with activity over a broad range of pHs and their overproduction by Pichia pastoris". Applied and Environmental Microbiology. 76 (19): 6423–30. doi:10.1128/AEM.00253-10. PMC 2950461. PMID 20693453.
- Konietzny U, Greiner R (2002). "Molecular and catalytic properties of phytate-degrading enzymes (phytases)". International Journal of Food Science and Technology. 37 (7): 791–812. doi:10.1046/j.1365-2621.2002.00617.x.
- Huang H, Shao N, Wang Y, Luo H, Yang P, Zhou Z, Zhan Z, Yao B (May 2009). "A novel beta-propeller phytase from Pedobacter nyackensis MJ11 CGMCC 2503 with potential as an aquatic feed additive". Applied Microbiology and Biotechnology. 83 (2): 249–59. doi:10.1007/s00253-008-1835-1. PMID 19139877.
- Döbereiner A, Schmid A, Ludwig A, Goebel W, Benz R (September 1996). "The effects of calcium and other polyvalent cations on channel formation by Escherichia coli alpha-hemolysin in red blood cells and lipid bilayer membranes". European Journal of Biochemistry. 240 (2): 454–60. doi:10.1111/j.1432-1033.1996.0454h.x. PMID 8841412.
- Kerovuo J, Rouvinen J, Hatzack F (December 2000). "Analysis of myo-inositol hexakisphosphate hydrolysis by Bacillus phytase: indication of a novel reaction mechanism". The Biochemical Journal. 352 Pt 3 (Pt 3): 623–8. PMC 1221497. PMID 11104666.
- Greiner R, Larsson Alminger M, Carlsson N, Muzquiz M, Burbano C, Cuadrado C, Pedrosa MM, Goyoaga C (2002). "Pathway of Dephosphorylation ofmyo-Inositol Hexakisphosphate by Phytases of Legume Seeds". Journal of Agricultural and Food Chemistry. 50 (23): 6865–6870. doi:10.1021/jf025620t.
- Oh BC, Kim MH, Yun BS, Choi WC, Park SC, Bae SC, Oh TK (August 2006). "Ca(2+)-inositol phosphate chelation mediates the substrate specificity of beta-propeller phytase". Biochemistry. 45 (31): 9531–9. doi:10.1021/bi0603118. PMID 16878987.
- Nam SJ, Kim YO, Ko TK, Kang JK, Chun KH, Auh JH, Lee CS, Lee IK, Park S, Oh BC (October 2014). "Molecular and biochemical characteristics of β-propeller phytase from marine Pseudomonas sp. BS10-3 and its potential application for animal feed additives". Journal of Microbiology and Biotechnology. 24 (10): 1413–20. doi:10.4014/jmb.1407.07063. PMID 25112322.